Reprogramming to a muscle fate by fusion recapitulates differentiation
- PMID: 19295131
- PMCID: PMC2720934
- DOI: 10.1242/jcs.041376
Reprogramming to a muscle fate by fusion recapitulates differentiation
Abstract
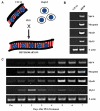
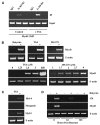
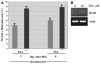
Fusion of mammalian cells to form stable, non-dividing heterokaryons results in nuclear reprogramming without the exchange of genetic material. In this report, we show that reprogramming in somatic cell heterokaryons involves activation of the canonical skeletal muscle transcription factors as well as contraction-excitation genes. Thus, the effect of heterokaryon formation on gene expression is to induce a recapitulation of differentiation. Heterokaryons formed with a relatively refractory cell type, the hepatocyte cell line HepG2, revealed the importance of both MyoD expression and other unidentified cytoplasmic components, neither of which are sufficient for efficient muscle gene activation, but are synergistic. We provide evidence that de-repression by transient histone deacetylase inhibition can induce MyoD expression and increase the extent and efficiency of muscle gene transcription. Taken together, the results suggest that understanding the mechanistic basis, using a combination of approaches, and taking into account cell history, will facilitate an increase in the efficiency and fidelity of conversion from one differentiated phenotype to another desired cell type. Inherent advantages of the heterokaryon system merit further investigation in the pursuit of directed cloning.
Figures

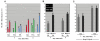

References
-
- Aoi, T., Yae, K., Nakagawa, M., Ichisaka, T., Okita, K., Takahashi, K., Chiba, T. and Yamanaka, S. (2008). Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321, 699-702. - PubMed
-
- Blakely, B. T. (1993). Negative and positive regulatory mechanisms in muscle and liver differentiation. PhD Thesis, Stanford University, Palo Alto, CA, USA.
-
- Blau, H. M., Chiu, C. P. and Webster, C. (1983). Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32, 1171-1180. - PubMed
-
- Blau, H. M., Chiu, C. P., Pavlath, G. K. and Webster, C. (1985a). Muscle gene expression in heterokaryons. Adv. Exp. Med. Biol. 182, 231-247. - PubMed
-
- Blau, H. M., Pavlath, G. K., Hardeman, E. C., Chiu, C. P., Silberstein, L., Webster, S. G., Miller, S. C. and Webster, C. (1985b). Plasticity of the differentiated state. Science 230, 758-766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

