Current tools for the identification of miRNA genes and their targets
- PMID: 19295136
- PMCID: PMC2677885
- DOI: 10.1093/nar/gkp145
Current tools for the identification of miRNA genes and their targets
Abstract
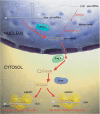
The discovery of microRNAs (miRNAs), almost 10 years ago, changed dramatically our perspective on eukaryotic gene expression regulation. However, the broad and important functions of these regulators are only now becoming apparent. The expansion of our catalogue of miRNA genes and the identification of the genes they regulate owe much to the development of sophisticated computational tools that have helped either to focus or interpret experimental assays. In this article, we review the methods for miRNA gene finding and target identification that have been proposed in the last few years. We identify some problems that current approaches have not yet been able to overcome and we offer some perspectives on the next generation of computational methods.
Figures
References
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. - PubMed
-
- Lai EC. microRNAs: runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. - PubMed
-
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

