Extended IL10 haplotypes and their association with HIV progression to AIDS
- PMID: 19295541
- PMCID: PMC3664918
- DOI: 10.1038/gene.2009.9
Extended IL10 haplotypes and their association with HIV progression to AIDS
Abstract
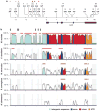
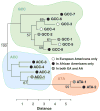
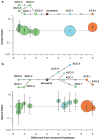
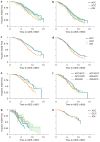
Interleukin-10 (IL-10) is a pleiotropic cytokine with both immunosuppressive and immunostimulatory functions. Its roles in infections and autoimmunity may have resulted in selective pressures on polymorphisms within the gene, leading to genomic coexistence of several semi-conserved haplotypes involved with diverse pathogen interactions during genomic evolution. Previous studies focused either exclusively on promoter haplotypes or on individual SNPs. We genotyped 21 single nucleotide polymorphisms in the human IL10 gene and examined this variation compared to other mammalian species sequences. Haplotype heterogeneity in human populations is centered around 'classic' 'proximal' promoter polymorphisms: -592, -819 and -1082. High-producing GCC haplotypes are by far the most numerous and diverse group, the intermediate IL-10 producing ACC-inclusive haplotypes seem to be related most closely to the ancestral haplotype, and the ATA-inclusive haplotypes cluster a separate branch with strong bootstrap support. We looked at associations of corresponding haplotypes with HIV progression. A haplotype trend regression confirmed that individuals carrying the low-producing ATA-inclusive haplotypes in European Americans progress to AIDS faster, and most likely explain the role of IL10. Our findings are consistent with the hypothesis that existing polymorphisms in this gene may reflect a balance of historic adaptive responses to autoimmune, infectious and other disease agents.
Figures
Similar articles
-
Frequency distribution of interleukin-10 haplotypes (-1082 A>G, -819 C>T, and -592 C>A) in a Mexican population.Genet Mol Res. 2016 Nov 3;15(4). doi: 10.4238/gmr15048411. Genet Mol Res. 2016. PMID: 27819740
-
Short Communication: Genetic Variation in Human IL10 Proximal Promoter and Susceptibility to HIV-1 Infection in Mali, West Africa.AIDS Res Hum Retroviruses. 2021 Jan;37(1):57-61. doi: 10.1089/AID.2020.0140. Epub 2020 Nov 23. AIDS Res Hum Retroviruses. 2021. PMID: 33045845 Free PMC article.
-
Interleukin-10 gene polymorphisms influence susceptibility to cachexia in patients with low-third gastric cancer in a Chinese population.Mol Diagn Ther. 2010 Apr 1;14(2):95-100. doi: 10.1007/BF03256358. Mol Diagn Ther. 2010. PMID: 20359252
-
Frequencies of IL10 SNP genotypes by multiplex PCR-SSP and their association with viral load and CD4 counts in HIV-1-infected Thais.Asian Pac J Allergy Immunol. 2011 Mar;29(1):94-101. Asian Pac J Allergy Immunol. 2011. PMID: 21560494
-
Associations Between Interleukin-10 Polymorphisms and Susceptibility to Sjögren's Syndrome: A Meta-Analysis.Int J Immunogenet. 2025 Feb;52(1):24-32. doi: 10.1111/iji.12702. Epub 2024 Nov 7. Int J Immunogenet. 2025. PMID: 39511968 Review.
Cited by
-
Polymorphisms in the IFNγ, IL-10, and TGFβ genes may be associated with HIV-1 infection.Dis Markers. 2015;2015:248571. doi: 10.1155/2015/248571. Epub 2015 Feb 24. Dis Markers. 2015. PMID: 25802474 Free PMC article.
-
Pilot study: possible association of IL10 promoter polymorphisms with CRMO.Rheumatol Int. 2012 Feb;32(2):555-6. doi: 10.1007/s00296-010-1768-8. Epub 2011 Jan 15. Rheumatol Int. 2012. PMID: 21240493 No abstract available.
-
Effect of host genetics on the development of cytomegalovirus retinitis in patients with AIDS.J Infect Dis. 2010 Aug 15;202(4):606-13. doi: 10.1086/654814. J Infect Dis. 2010. PMID: 20617924 Free PMC article.
-
Influence of cytokines on HIV-specific antibody-dependent cellular cytotoxicity activation profile of natural killer cells.PLoS One. 2012;7(6):e38580. doi: 10.1371/journal.pone.0038580. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701674 Free PMC article.
-
The Enigmatic Interplay of Interleukin-10 in the Synergy of HIV Infection Comorbid with Preeclampsia.Int J Mol Sci. 2024 Aug 30;25(17):9434. doi: 10.3390/ijms25179434. Int J Mol Sci. 2024. PMID: 39273381 Free PMC article. Review.
References
-
- UNAIDS. AIDS Epidemic Update. UNAIDS/WHO; Geneva, Switzerland: 2006.
-
- Dumont F. Therapeutic potential of IL-10 and its viral homologues: an update. Expert Opin Ther Patents. 2003;13:1551–1577.
-
- O’Brien TR, Winkler C, Dean M, Nelson JA, Carrington M, Michael NL, et al. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. - PubMed
-
- Tang J, Penman-Aguilar A, Lobashevsky E, Allen S, Kaslow RA. HLA-DRB1 and -DQB1 alleles and haplotypes in Zambian couples and their associations with heterosexual transmission of HIV type 1. J Infect Dis. 2004;189:1696–1704. - PubMed
-
- Shrestha S, Strathdee SA, Galai N, Oleksyk TK, Fallin MD, Mehta S, et al. Behavioral risk exposure and host genetics of susceptibility to HIV-1 infection. J Infect Dis. 2006;193:16–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

