A computational approach for genome-wide mapping of splicing factor binding sites
- PMID: 19296853
- PMCID: PMC2691001
- DOI: 10.1186/gb-2009-10-3-r30
A computational approach for genome-wide mapping of splicing factor binding sites
Abstract
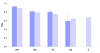
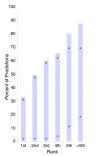
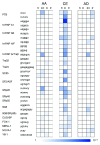
Alternative splicing is regulated by splicing factors that serve as positive or negative effectors, interacting with regulatory elements along exons and introns. Here we present a novel computational method for genome-wide mapping of splicing factor binding sites that considers both the genomic environment and the evolutionary conservation of the regulatory elements. The method was applied to study the regulation of different alternative splicing events, uncovering an interesting network of interactions among splicing factors.
Figures
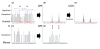


References
-
- Das D, Clark TA, Schweitzer A, Yamamoto M, Marr H, Arribere J, Minovitsky S, Poliakov A, Dubchak I, Blume JE, Conboy JG. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 2007;35:4845–4857. doi: 10.1093/nar/gkm485. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

