Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers
- PMID: 19297317
- PMCID: PMC2679481
- DOI: 10.1074/jbc.M901351200
Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers
Abstract
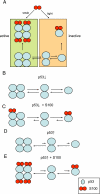
We investigated the ways S100B, S100A1, S100A2, S100A4, and S100A6 bind to the different oligomeric forms of the tumor suppressor p53 in vitro, using analytical ultracentrifugation and multiangle light scattering. It is established that members of the S100 protein family bind to the tetramerization domain (residues 325-355) of p53 when it is uncovered in the monomer, and so binding can disrupt the tetramer. We found a stoichiometry of one dimer of S100 bound to a monomer of p53. We discovered that some S100 proteins could also bind to the tetramer. S100B bound the tetramer and also disrupted the dimer by binding monomeric p53. S100A2 bound monomeric p53 as well as tetrameric, whereas S100A1 only bound monomeric p53. S100A6 bound more tightly to tetrameric than to monomeric p53. We also identified an additional binding site for S100 proteins in the transactivation domain (1-57) of p53. Based on our results and published observations in vivo, we propose a model for the binding of S100 proteins to p53 that can explain both activation and inhibition of p53-mediated transcription. Depending on the concentration of p53 and the member of the S100 family, binding can alter the balance between monomer and tetramer in either direction.
Figures
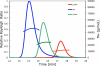
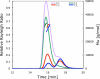
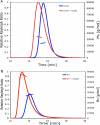

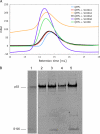

References
-
- Donato, R. (2003) Microsc. Res. Tech. 60 540-551 - PubMed
-
- Moore, B. W. (1965) Biochem. Biophys. Res. Commun. 19 739-744 - PubMed
-
- Koch, M., Bhattacharya, S., Kehl, T., Gimona, M., Vasak, M., Chazin, W., Heizmann, C. W., Kroneck, P. M., and Fritz, G. (2007) Biochim. Biophys. Acta 1773 457-470 - PubMed
-
- Donato, R. (2001) Int. J. Biochem. Cell Biol. 33 637-668 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

