Structural insight into the activation mechanism of human pancreatic prophospholipase A2
- PMID: 19297324
- PMCID: PMC2713533
- DOI: 10.1074/jbc.M808029200
Structural insight into the activation mechanism of human pancreatic prophospholipase A2
Abstract
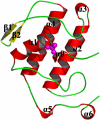
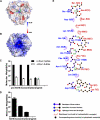
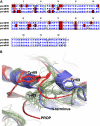
Pancreatic phospholipase A2 (phospholipase A2 group 1B, G1B) belongs to the superfamily of secreted phospholipase A2 (PLA2) enzymes. G1B has been proposed to be a potential target for diseases such as hypertension, obesity, and diabetes. Human pancreatic prophospholipase A2 (pro-hG1B) is activated by cleavage of the first seven-residue propeptide (phospholipase A2 propeptide, PROP). However, questions still remain on the mode of action for pro-hG1B. In this work, we expressed pro-hG1B in Pichia pastoris and determined the crystal structure at 1.55-A resolution. The x-ray structure demonstrates that pro-hG1B forms a trimer. In addition, PROP occupies the catalytic cavity and can be self-cleaved at 37 degrees C. A new membrane-bound surface and activation mechanism are proposed based on the trimeric model of pro-hG1B. We also propose a new autoproteolytic mechanism for pro-hG1B by the reaction triad Asp49-Arg0-Ser(-2) that is similar to the serine protease catalytic triad.
Figures
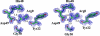

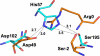
References
-
- Schaloske R. H., Dennis E. A. ( 2006) Biochim. Biophys. Acta 1761, 1246– 1259 - PubMed
-
- Frossard P. M., Lestringant G. G. ( 1995) Clin. Genet. 48, 284– 287 - PubMed
-
- Kuopio T., Ekfors T. O., Nikkanen V., Nevalainen T. J. ( 1995) Apmis. 103, 69– 78 - PubMed
-
- Huggins K. W., Boileau A. C., Hui D. Y. ( 2002) Am. J. Physiol. Endocrinol. Metab. 283, E994– 1001 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

