Cell-to-cell spread of the RNA interference response suppresses Semliki Forest virus (SFV) infection of mosquito cell cultures and cannot be antagonized by SFV
- PMID: 19297476
- PMCID: PMC2681975
- DOI: 10.1128/JVI.02440-08
Cell-to-cell spread of the RNA interference response suppresses Semliki Forest virus (SFV) infection of mosquito cell cultures and cannot be antagonized by SFV
Abstract
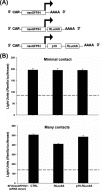
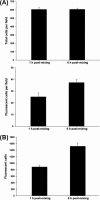
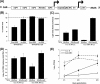
In their vertebrate hosts, arboviruses such as Semliki Forest virus (SFV) (Togaviridae) generally counteract innate defenses and trigger cell death. In contrast, in mosquito cells, following an early phase of efficient virus production, a persistent infection with low levels of virus production is established. Whether arboviruses counteract RNA interference (RNAi), which provides an important antiviral defense system in mosquitoes, is an important question. Here we show that in Aedes albopictus-derived mosquito cells, SFV cannot prevent the establishment of an antiviral RNAi response or prevent the spread of protective antiviral double-stranded RNA/small interfering RNA (siRNA) from cell to cell, which can inhibit the replication of incoming virus. The expression of tombusvirus siRNA-binding protein p19 by SFV strongly enhanced virus spread between cultured cells rather than virus replication in initially infected cells. Our results indicate that the spread of the RNAi signal contributes to limiting virus dissemination.
Figures
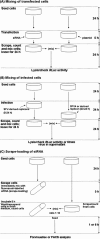
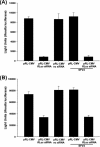
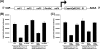
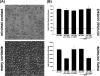



References
-
- Adelman, Z. N., I. Sanchez-Vargas, E. A. Travanty, J. O. Carlson, B. J. Beaty, C. D. Blair, and K. E. Olson. 2002. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 7612925-12933. - PMC - PubMed
-
- Baulcombe, D. 2004. RNA silencing in plants. Nature 431356-363. - PubMed
-
- Billecocq, A., M. Vazeille-Falcoz, F. Rodhain, and M. Bouloy. 2000. Pathogen-specific resistance to Rift Valley fever virus infection is induced in mosquito cells by expression of the recombinant nucleoprotein but not NSs non-structural protein sequences. J. Gen. Virol. 812161-2166. - PubMed
-
- Blakqori, G., S. Delhaye, M. Habjan, C. D. Blair, I. Sanchez-Vargas, K. E. Olson, G. Attarzadeh-Yazdi, R. Fragkoudis, A. Kohl, U. Kalinke, S. Weiss, T. Michiels, P. Staeheli, and F. Weber. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 814991-4999. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

