Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters
- PMID: 19297587
- PMCID: PMC2675721
- DOI: 10.1104/pp.109.135624
Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters
Abstract
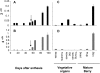
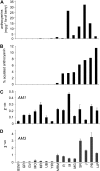
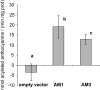
In grapevine (Vitis vinifera), anthocyanins are responsible for most of the red, blue, and purple pigmentation found in the skin of berries. In cells, anthocyanins are synthesized in the cytoplasm and accumulated into the vacuole. However, little is known about the transport of these compounds through the tonoplast. Recently, the sequencing of the grapevine genome allowed us to identify genes encoding proteins with high sequence similarity to the Multidrug And Toxic Extrusion (MATE) family. Among them, we selected two genes as anthocyanin transporter candidates and named them anthoMATE1 (AM1) and AM3. The expression of both genes was mainly fruit specific and concomitant with the accumulation of anthocyanin pigment. Subcellular localization assays in grapevine hairy roots stably transformed with AM1 or AM3green fluorescent protein fusion protein revealed that AM1 and AM3 are primarily localized to the tonoplast. Yeast vesicles expressing anthoMATEs transported acylated anthocyanins in the presence of MgATP. Inhibitor studies demonstrated that AM1 and AM3 proteins act in vitro as vacuolar H(+)-dependent acylated anthocyanin transporters. By contrast, under our experimental conditions, anthoMATEs could not transport malvidin 3-O-glucoside or cyanidin 3-O-glucoside, suggesting that the acyl conjugation was essential for the uptake. Taken together, these results provide evidence that in vitro the two grapevine AM1 and AM3 proteins mediate specifically acylated anthocyanin transport.
Figures





References
-
- Ageorges A, Fernandez L, Vialet S, Merdinoglu D, Terrier N, Romieu C (2006) Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci 170 372–383
-
- Archetti M (2000) The origin of autumn colours by coevolution. J Theor Biol 205 625–630 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

