A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments
- PMID: 19298048
- PMCID: PMC2693032
- DOI: 10.1021/jp810375d
A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments
Abstract
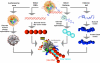
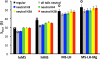
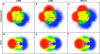
To elucidate the role of the histone tails in chromatin compaction and in higher-order folding of chromatin under physiological conditions, we extend a mesoscale model of chromatin (Arya, Zhang, and Schlick. Biophys. J. 2006, 91, 133; Arya and Schlick. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 16236) to account for divalent cations (Mg(2+)) and linker histones. Configurations of 24-nucleosome oligonucleosomes in different salt environments and in the presence and absence of linker histones are sampled by a mixture of local and global Monte Carlo methods. Analyses of the resulting ensembles reveal a dynamic synergism between the histone tails, linker histones, and ions in forming compact higher-order structures of chromatin. In the presence of monovalent salt alone, oligonucleosomes remain relatively unfolded, and the histone tails do not mediate many internucleosomal interactions. Upon the addition of linker histones and divalent cations, the oligonucleosomes undergo a significant compaction triggered by a dramatic increase in the internucleosomal interactions mediated by the histone tails, formation of a rigid linker DNA "stem" around the linker histones' C-terminal domains, and reduction in the electrostatic repulsion between linker DNAs via sharp bending in some linker DNAs caused by the divalent cations. Among all histone tails, the H4 tails mediate the most internucleosomal interactions, consistent with experimental observations, followed by the H3, H2A, and H2B tails in decreasing order. Apart from mediating internucleosomal interactions, the H3 tails also contribute to chromatin compaction by attaching to the entering and exiting linker DNA to screen electrotatic repulsion among the linker DNAs. This tendency of the H3 tails to attach to linker DNA, however, decreases significantly upon the addition of linker histones due to competition effects. The H2A and H2B tails do not mediate significant internucleosomal interactions but are important for mediating fiber/fiber intractions, especially in relatively unfolded chromatin in monovalent salt environments.
Figures




References
-
- Felsenfeld G, Groudine M. Nature. 2003;421:448–453. - PubMed
-
- Horn PJ, Peterson CL. Science. 2002;297:1824–1827. - PubMed
-
- Tse C, Hansen JC. Biochemistry. 1997;36:11381–11388. - PubMed
-
- Hansen JC, Tse C, Wolffe AP. Biochemistry. 1998;37:17637–17641. - PubMed
-
- Gordon F, Luger K, Hansen JC. J. Biol. Chem. 2005;280:33701–3706. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

