A comprehensive analysis of the La-motif protein superfamily
- PMID: 19299548
- PMCID: PMC2673062
- DOI: 10.1261/rna.1478709
A comprehensive analysis of the La-motif protein superfamily
Abstract
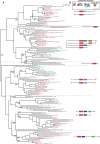
The extremely well-conserved La motif (LAM), in synergy with the immediately following RNA recognition motif (RRM), allows direct binding of the (genuine) La autoantigen to RNA polymerase III primary transcripts. This motif is not only found on La homologs, but also on La-related proteins (LARPs) of unrelated function. LARPs are widely found amongst eukaryotes and, although poorly characterized, appear to be RNA-binding proteins fulfilling crucial cellular functions. We searched the fully sequenced genomes of 83 eukaryotic species scattered along the tree of life for the presence of LAM-containing proteins. We observed that these proteins are absent from archaea and present in all eukaryotes (except protists from the Plasmodium genus), strongly suggesting that the LAM is an ancestral motif that emerged early after the archaea-eukarya radiation. A complete evolutionary and structural analysis of these proteins resulted in their classification into five families: the genuine La homologs and four LARP families. Unexpectedly, in each family a conserved domain representing either a classical RRM or an RRM-like motif immediately follows the LAM of most proteins. An evolutionary analysis of the LAM-RRM/RRM-L regions shows that these motifs co-evolved and should be used as a single entity to define the functional region of interaction of LARPs with their substrates. We also found two extremely well conserved motifs, named LSA and DM15, shared by LARP6 and LARP1 family members, respectively. We suggest that members of the same family are functional homologs and/or share a common molecular mode of action on different RNA baits.
Figures





References
-
- Adilakshmi T., Laine R.O. Ribosomal protein S25 mRNA partners with MTF-1 and La to provide a p53-mediated mechanism for survival or death. J. Biol. Chem. 2002;277:4147–4151. - PubMed
-
- Aigner S., Postberg J., Lipps H.J., Cech T.R. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry. 2003;42:5736–5747. - PubMed
-
- Alfano C., Sanfelice D., Babon J., Kelly G., Jacks A., Curry S., Conte M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004;11:323–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
