Parallel germline infiltration of a lentivirus in two Malagasy lemurs
- PMID: 19300488
- PMCID: PMC2651035
- DOI: 10.1371/journal.pgen.1000425
Parallel germline infiltration of a lentivirus in two Malagasy lemurs
Abstract
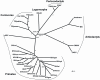
Retroviruses normally infect the somatic cells of their host and are transmitted horizontally, i.e., in an exogenous way. Occasionally, however, some retroviruses can also infect and integrate into the genome of germ cells, which may allow for their vertical inheritance and fixation in a given species; a process known as endogenization. Lentiviruses, a group of mammalian retroviruses that includes HIV, are known to infect primates, ruminants, horses, and cats. Unlike many other retroviruses, these viruses have not been demonstrably successful at germline infiltration. Here, we report on the discovery of endogenous lentiviral insertions in seven species of Malagasy lemurs from two different genera -- Cheirogaleus and Microcebus. Combining molecular clock analyses and cross-species screening of orthologous insertions, we show that the presence of this endogenous lentivirus in six species of Microcebus is the result of one endogenization event that occurred about 4.2 million years ago. In addition, we demonstrate that this lentivirus independently infiltrated the germline of Cheirogaleus and that the two endogenization events occurred quasi-simultaneously. Using multiple proviral copies, we derive and characterize an apparently full length and intact consensus for this lentivirus. These results provide evidence that lentiviruses have repeatedly infiltrated the germline of prosimian species and that primates have been exposed to lentiviruses for a much longer time than what can be inferred based on sequence comparison of circulating lentiviruses. The study sets the stage for an unprecedented opportunity to reconstruct an ancestral primate lentivirus and thereby advance our knowledge of host-virus interactions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
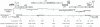




Similar articles
-
A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution.Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20362-7. doi: 10.1073/pnas.0807873105. Epub 2008 Dec 15. Proc Natl Acad Sci U S A. 2008. PMID: 19075221 Free PMC article.
-
A primitive endogenous lentivirus in a colugo: insights into the early evolution of lentiviruses.Mol Biol Evol. 2015 Jan;32(1):211-5. doi: 10.1093/molbev/msu297. Epub 2014 Oct 27. Mol Biol Evol. 2015. PMID: 25349288 Free PMC article.
-
Endogenization of a Prosimian Retrovirus during Lemur Evolution.Viruses. 2021 Feb 27;13(3):383. doi: 10.3390/v13030383. Viruses. 2021. PMID: 33673677 Free PMC article.
-
Transmission, Evolution, and Endogenization: Lessons Learned from Recent Retroviral Invasions.Microbiol Mol Biol Rev. 2017 Dec 13;82(1):e00044-17. doi: 10.1128/MMBR.00044-17. Print 2018 Mar. Microbiol Mol Biol Rev. 2017. PMID: 29237726 Free PMC article. Review.
-
Human endogenous retroviruses in the primate lineage and their influence on host genomes.Cytogenet Genome Res. 2005;110(1-4):448-56. doi: 10.1159/000084977. Cytogenet Genome Res. 2005. PMID: 16093697 Review.
Cited by
-
Species-specific transmission of novel picornaviruses in lemurs.J Virol. 2015 Apr;89(7):4002-10. doi: 10.1128/JVI.03342-14. Epub 2015 Jan 28. J Virol. 2015. PMID: 25631076 Free PMC article.
-
Pan-vertebrate comparative genomics unmasks retrovirus macroevolution.Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):464-9. doi: 10.1073/pnas.1414980112. Epub 2014 Dec 22. Proc Natl Acad Sci U S A. 2015. PMID: 25535393 Free PMC article.
-
Betaretroviral envelope subunits are noncovalently associated and restricted to the mammalian class.J Virol. 2013 Feb;87(4):1937-46. doi: 10.1128/JVI.01442-12. Epub 2012 Dec 5. J Virol. 2013. PMID: 23221553 Free PMC article.
-
Cyclophilin A protects HIV-1 from restriction by human TRIM5α.Nat Microbiol. 2019 Dec;4(12):2044-2051. doi: 10.1038/s41564-019-0592-5. Epub 2019 Oct 21. Nat Microbiol. 2019. PMID: 31636416 Free PMC article.
-
The diversity and evolution of retroviruses: Perspectives from viral "fossils".Virol Sin. 2022 Feb;37(1):11-18. doi: 10.1016/j.virs.2022.01.019. Epub 2022 Jan 19. Virol Sin. 2022. PMID: 35234634 Free PMC article. Review.
References
-
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–14. - PubMed
-
- Gifford RJ. Evolution at the host-retrovirus interface. Bioessays. 2006;28:1153–1156. - PubMed
-
- Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Press; 1997. 843 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

