Genetic drift of HIV populations in culture
- PMID: 19300501
- PMCID: PMC2652835
- DOI: 10.1371/journal.pgen.1000431
Genetic drift of HIV populations in culture
Abstract
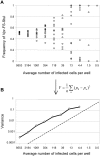
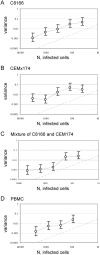
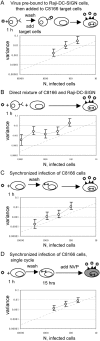
Populations of Human Immunodeficiency Virus type 1 (HIV-1) undergo a surprisingly large amount of genetic drift in infected patients despite very large population sizes, which are predicted to be mostly deterministic. Several models have been proposed to explain this phenomenon, but all of them implicitly assume that the process of virus replication itself does not contribute to genetic drift. We developed an assay to measure the amount of genetic drift for HIV populations replicating in cell culture. The assay relies on creation of HIV populations of known size and measurements of variation in frequency of a neutral allele. Using this assay, we show that HIV undergoes approximately ten times more genetic drift than would be expected from its population size, which we defined as the number of infected cells in the culture. We showed that a large portion of the increase in genetic drift is due to non-synchronous infection of target cells. When infections are synchronized, genetic drift for the virus is only 3-fold higher than expected from its population size. Thus, the stochastic nature of biological processes involved in viral replication contributes to increased genetic drift in HIV populations. We propose that appreciation of these effects will allow better understanding of the evolutionary forces acting on HIV in infected patients.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- French R, Stenger DC. Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu Rev Phytopathol. 2003;41:199–214. - PubMed
-
- French R, Stenger DC. Population structure within lineages of Wheat streak mosaic virus derived from a common founding event exhibits stochastic variation inconsistent with the deterministic quasi-species model. Virology. 2005;343:179–189. - PubMed
-
- Hall G. Selective constraint and genetic differentiation in geographically distant barley yellow dwarf virus populations. J Gen Virol. 2006;87:3067–3075. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

