Emergent genome-wide control in wildtype and genetically mutated lipopolysaccarides-stimulated macrophages
- PMID: 19300509
- PMCID: PMC2654147
- DOI: 10.1371/journal.pone.0004905
Emergent genome-wide control in wildtype and genetically mutated lipopolysaccarides-stimulated macrophages
Abstract
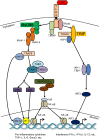
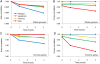
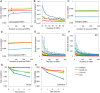
Large-scale gene expression studies have mainly focused on highly expressed and 'discriminatory' genes to decipher key regulatory processes. Biological responses are consequence of the concerted action of gene regulatory network, thus, limiting our attention to genes having the most significant variations is insufficient for a thorough understanding of emergent whole genome response. Here we comprehensively analyzed the temporal oligonucleotide microarray data of lipopolysaccharide (LPS) stimulated macrophages in 4 genotypes; wildtype, Myeloid Differentiation factor 88 (MyD88) knockout (KO), TIR-domain-containing adapter-inducing interferon-beta (TRIF) KO and MyD88/TRIF double KO (DKO). Pearson correlations computed on the whole genome expression between different genotypes are extremely high (>0.98), indicating a strong co-regulation of the entire expression network. Further correlation analyses reveal genome-wide response is biphasic, i) acute-stochastic mode consisting of small number of sharply induced immune-related genes and ii) collective mode consisting of majority of weakly induced genes of diverse cellular processes which collectively adjust their expression level. Notably, temporal correlations of a small number of randomly selected genes from collective mode show scalability. Furthermore, in collective mode, the transition from large scatter in expression distributions for single ORFs to smooth linear lines emerges as an organizing principle when grouping of 50 ORFs and above. With this emergent behavior, the role of MyD88, TRIF and novel MyD88, TRIF-independent processes for gene induction can be linearly superposed to decipher quantitative whole genome differential control of transcriptional and mRNA decay machineries. Our work demonstrates genome-wide co-regulated responses subsequent to specific innate immune stimulus which have been largely neglected.
Conflict of interest statement
Figures



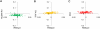
References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. - PubMed
-
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. - PubMed
-
- Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

