The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans
- PMID: 19300908
- PMCID: PMC11115746
- DOI: 10.1007/s00018-009-0010-x
The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans
Abstract
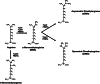
Information about the family of protein arginine methyltransferases (PRMTs) has been growing rapidly over the last few years and the emerging role of arginine methylation involved in cellular processes like signaling, RNA processing, gene transcription, and cellular transport function has been investigated. To date, 11 PRMTs gene transcripts have been identified in humans. Almost all PRMTs have been shown to have enzymatic activity and to catalyze arginine methylation. This review will summarize the overall function of human PRMTs and include novel highlights on each family member.
Figures

Similar articles
-
Toward Understanding Molecular Recognition between PRMTs and their Substrates.Curr Protein Pept Sci. 2020;21(7):713-724. doi: 10.2174/1389203721666200124143145. Curr Protein Pept Sci. 2020. PMID: 31976831 Review.
-
Protein arginine methylation: Cellular functions and methods of analysis.Biochim Biophys Acta. 2006 Dec;1764(12):1890-903. doi: 10.1016/j.bbapap.2006.08.008. Epub 2006 Aug 25. Biochim Biophys Acta. 2006. PMID: 17010682 Review.
-
Non-Histone Arginine Methylation by Protein Arginine Methyltransferases.Curr Protein Pept Sci. 2020;21(7):699-712. doi: 10.2174/1389203721666200507091952. Curr Protein Pept Sci. 2020. PMID: 32379587 Free PMC article. Review.
-
The regulation, functions and clinical relevance of arginine methylation.Nat Rev Mol Cell Biol. 2019 Oct;20(10):642-657. doi: 10.1038/s41580-019-0155-x. Epub 2019 Jul 26. Nat Rev Mol Cell Biol. 2019. PMID: 31350521 Review.
-
Arginine methylation and respiratory disease.Transl Res. 2024 Oct;272:140-150. doi: 10.1016/j.trsl.2024.03.002. Epub 2024 Mar 5. Transl Res. 2024. PMID: 38453053 Review.
Cited by
-
Protein arginine methyltransferase 5 is essential for growth of lung cancer cells.Biochem J. 2012 Sep 1;446(2):235-41. doi: 10.1042/BJ20120768. Biochem J. 2012. PMID: 22708516 Free PMC article.
-
Protein arginine methyltransferase Prmt5-Mep50 methylates histones H2A and H4 and the histone chaperone nucleoplasmin in Xenopus laevis eggs.J Biol Chem. 2011 Dec 9;286(49):42221-42231. doi: 10.1074/jbc.M111.303677. Epub 2011 Oct 18. J Biol Chem. 2011. PMID: 22009756 Free PMC article.
-
Histones and heart failure in diabetes.Cell Mol Life Sci. 2018 Sep;75(17):3193-3213. doi: 10.1007/s00018-018-2857-1. Epub 2018 Jun 22. Cell Mol Life Sci. 2018. PMID: 29934664 Free PMC article. Review.
-
Prmt7 is required for the osteogenic differentiation of mesenchymal stem cells via modulation of BMP signaling.BMB Rep. 2024 Jul;57(7):330-335. doi: 10.5483/BMBRep.2023-0203. BMB Rep. 2024. PMID: 38627951 Free PMC article.
-
Crosstalk between metabolism and epigenetics during macrophage polarization.Epigenetics Chromatin. 2025 Mar 29;18(1):16. doi: 10.1186/s13072-025-00575-9. Epigenetics Chromatin. 2025. PMID: 40156046 Free PMC article. Review.
References
-
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. - PubMed
-
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. - PubMed
-
- Lee DJ, Teyssier C, Strahl BD, Stallcup MR. Role of methylation in regulation of transcription. Endocr Rev. 2005;26:147–170. - PubMed
-
- Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

