Secreted TARSH regulates olfactory mitral cell dendritic complexity
- PMID: 19302145
- PMCID: PMC2772113
- DOI: 10.1111/j.1460-9568.2009.06660.x
Secreted TARSH regulates olfactory mitral cell dendritic complexity
Abstract
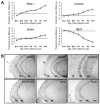
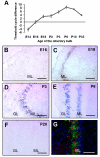
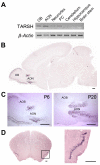
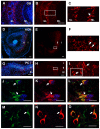
Olfactory sensory neurons synapse with mitral cells to form stereotyped connections in the olfactory bulb (OB). Mitral cell apical dendrites receive input from olfactory sensory neurons expressing the same odorant receptor. During development, this restricted dendritic targeting of mitral cells is achieved through eliminating elaborated dendritic trees to a single apical dendrite. Through a genome-wide microarray screen, we identified TARSH (Target of NESH SH3) as a transiently expressed molecule in mitral cells during the dendritic refinement period. TARSH expression is restricted to pyramidal neurons along the main olfactory pathway, including the anterior olfactory nucleus and piriform cortex. The dynamic TARSH expression is not altered when odor-evoked activity is blocked by naris closure or in AC3 knockout mice. We also demonstrate that TARSH is a secreted protein. In dissociated OB cultures, secreted TARSH promotes the reduction of mitral cell dendritic complexity and restricts dendritic branching and outgrowth of interneurons. Dendritic morphological changes were also observed in mitral cells overexpressing TARSH themselves. We propose that TARSH is part of the genetic program that regulates mitral cell dendritic refinement.
Figures
Similar articles
-
Nectin-1 spots regulate the branching of olfactory mitral cell dendrites.Mol Cell Neurosci. 2015 Sep;68:143-50. doi: 10.1016/j.mcn.2015.07.003. Epub 2015 Jul 11. Mol Cell Neurosci. 2015. PMID: 26169026
-
5T4 glycoprotein regulates the sensory input-dependent development of a specific subtype of newborn interneurons in the mouse olfactory bulb.J Neurosci. 2012 Feb 8;32(6):2217-26. doi: 10.1523/JNEUROSCI.5907-11.2012. J Neurosci. 2012. PMID: 22323733 Free PMC article.
-
Dendritic branching of olfactory bulb mitral and tufted cells: regulation by TrkB.PLoS One. 2009 Aug 25;4(8):e6729. doi: 10.1371/journal.pone.0006729. PLoS One. 2009. PMID: 19707543 Free PMC article.
-
Dendritic processing within olfactory bulb circuits.Trends Neurosci. 2003 Sep;26(9):501-6. doi: 10.1016/S0166-2236(03)00228-5. Trends Neurosci. 2003. PMID: 12948662 Review.
-
Odor representations in mammalian cortical circuits.Curr Opin Neurobiol. 2010 Jun;20(3):328-31. doi: 10.1016/j.conb.2010.02.004. Epub 2010 Mar 5. Curr Opin Neurobiol. 2010. PMID: 20207132 Free PMC article. Review.
Cited by
-
Complexin activates exocytosis of distinct secretory vesicles controlled by different synaptotagmins.J Neurosci. 2013 Jan 23;33(4):1714-27. doi: 10.1523/JNEUROSCI.4087-12.2013. J Neurosci. 2013. PMID: 23345244 Free PMC article.
-
Secretome survey of human plexiform neurofibroma derived Schwann Cells reveals a secreted form of the RARRES1 protein.Int J Mol Sci. 2012;13(7):9380-9399. doi: 10.3390/ijms13079380. Epub 2012 Jul 24. Int J Mol Sci. 2012. PMID: 22942771 Free PMC article.
-
Abi3bp regulates cardiac progenitor cell proliferation and differentiation.Circ Res. 2014 Dec 5;115(12):1007-16. doi: 10.1161/CIRCRESAHA.115.304216. Epub 2014 Oct 8. Circ Res. 2014. PMID: 25296984 Free PMC article.
-
Abi3bp is a multifunctional autocrine/paracrine factor that regulates mesenchymal stem cell biology.Stem Cells. 2013 Aug;31(8):1669-82. doi: 10.1002/stem.1416. Stem Cells. 2013. PMID: 23666637 Free PMC article.
-
Study on the correlation between suicidal ideation and ABI3BP gene、DPYSL2 gene methylation in pediatric bipolar disorder with depressive episode.Heliyon. 2023 Dec 14;10(1):e23680. doi: 10.1016/j.heliyon.2023.e23680. eCollection 2024 Jan 15. Heliyon. 2023. PMID: 38226278 Free PMC article.
References
-
- Bagri A, Cheng HJ, Yaron A, Pleasure SJ, Tessier-Lavigne M. Stereotyped pruning of long hippocampal axon branches triggered by retraction inducers of the semaphorin family. Cell. 2003;113:285–299. - PubMed
-
- Bjartmar L, Huberman AD, Ullian EM, Renteria RC, Liu X, Xu W, Prezioso J, Susman MW, Stellwagen D, Stokes CC, Cho R, Worley P, Malenka RC, Ball S, Peachey NS, Copenhagen D, Chapman B, Nakamoto M, Barres BA, Perin MS. Neuronal pentraxins mediate synaptic refinement in the developing visual system. J Neurosci. 2006;26:6269–6281. - PMC - PubMed
-
- Blanchart A, De Carlos JA, Lopez-Mascaraque L. Time frame of mitral cell development in the mice olfactory bulb. J Comp Neurol. 2006;496:529–543. - PubMed
-
- Chen H, Kohno K, Gong Q. Conditional ablation of mature olfactory sensory neurons mediated by diphtheria toxin receptor. J Neurocytol. 2005;34:37–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

