Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein
- PMID: 19303045
- PMCID: PMC2680464
- DOI: 10.1016/j.gene.2009.02.022
Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein
Abstract
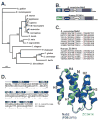
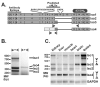
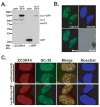
The human ZC3H14 gene encodes an evolutionarily conserved Cys(3)His zinc finger protein that binds specifically to polyadenosine RNA and is thus postulated to modulate post-transcriptional gene expression. Expressed sequence tag (EST) data predicts multiple splice variants of both human and mouse ZC3H14. Analysis of ZC3H14 expression in both human cell lines and mouse tissues confirms the presence of multiple alternatively spliced transcripts. Although all of these transcripts encode protein isoforms that contain the conserved C-terminal zinc finger domain, suggesting that they could all bind to polyadenosine RNA, they differ in other functionally important domains. Most of the alternative transcripts encode closely related proteins (termed isoforms 1, 2, 3, and 3 short) that differ primarily in the inclusion of three small exons, 9, 10, and 11, resulting in predicted protein isoforms ranging from 82 to 64 kDa. Each of these closely related isoforms contains predicted classical nuclear localization signals (cNLS) within exons 7 and 11. Consistent with the presence of these putative nuclear targeting signals, these ZC3H14 isoforms are all localized to the nucleus. In contrast, an additional transcript encodes a smaller protein (34 kDa) with an alternative first exon (isoform 4). Consistent with the absence of the predicted cNLS motifs located in exons 7 and 11, ZC3H14 isoform 4 is localized to the cytoplasm. Both EST data and experimental data suggest that this variant is enriched in testes and brain. Using an antibody that detects endogenous ZC3H14 isoforms 1-3 reveals localization of these isoforms to nuclear speckles. These speckles co-localize with the splicing factor, SC35, suggesting a role for nuclear ZC3H14 in mRNA processing. Taken together, these results demonstrate that multiple transcripts encoding several ZC3H14 isoforms exist in vivo. Both nuclear and cytoplasmic ZC3H14 isoforms could have distinct effects on gene expression mediated by the common Cys(3)His zinc finger polyadenosine RNA binding domain.
Figures

References
-
- Aitchison JD, Blobel G, Rout MP. Kap104p: A karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

