The long and the short of riboswitches
- PMID: 19303767
- PMCID: PMC2762789
- DOI: 10.1016/j.sbi.2009.02.002
The long and the short of riboswitches
Abstract
Regulatory mRNA elements or riboswitches specifically control the expression of a large number of genes in response to various cellular metabolites. The basis for selectivity of regulation is programmed in the evolutionarily conserved metabolite-sensing regions of riboswitches, which display a plethora of sequence and structural variants. Recent X-ray structures of two distinct SAM riboswitches and the sensing domains of the Mg(2+), lysine, and FMN riboswitches have uncovered novel recognition principles and provided molecular details underlying the exquisite specificity of metabolite binding by RNA. These and earlier structures constitute the majority of widespread riboswitch classes and, together with riboswitch folding studies, improve our understanding of the mechanistic principles involved in riboswitch-mediated gene expression control.
Figures
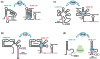

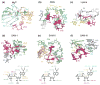
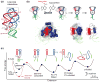
References
-
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. - PubMed
-
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. - PubMed
-
- Bocobza SE, Aharoni A. Switching the light on plant riboswitches. Trends Plant Sci. 2008;13:526–533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

