Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent
- PMID: 19304304
- PMCID: PMC3708462
- DOI: 10.1016/j.virol.2009.02.026
Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent
Abstract
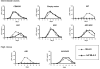
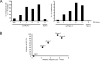
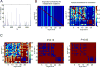
Cytidine deamination is the primary mechanism by which APOBEC3G restricts HIV-1; however, several studies have reported that APOBEC3G also inhibits virus replication via a mechanism that is independent of deamination. Using active site APOBEC3G mutants, we have re-evaluated the biological relevance of deaminase-independent APOBEC3G-mediated restriction of HIV-1. APOBEC3G proteins with Glu-->Ala mutations in AS1, AS2 or AS1 and AS2 were stably expressed at physiological levels in CEM-SS T cells and 293T cells and the ability of the cells to support Deltavif HIV-1 replication was then tested. The AS2 and AS1/AS2 mutants were packaged efficiently into virions but in single-cycle or multi-cycle HIV-1 replication assays, were found to lack antiviral activity. The AS1 mutant, which retained deaminase activity, maintained near wild-type antiviral function. To determine the potency of APOBEC3G antiviral activity, cell lines were established that that expressed low levels of wild-type APOBEC3G and generated virions that contained as few as 1-2 APOBEC3G molecules. Even at very low copy number, APOBEC3G caused a significant reduction in infectivity, suggesting that a single molecule of packaged APOBEC3G inactivates the virus. The high potency of APOBEC3G is consistent with a catalytic mechanism of restriction in which a single molecule can induce a string of mutations but difficult to reconcile with a deaminase-independent, non-catalytic mechanism. Analysis of the reverse transcript sequences showed that the G-->A mutations were clustered, likely reflecting the action of single APOBEC3G molecules acting processively. We conclude that cytidine deamination is the mechanism by which APOBEC3G restricts HIV-1.
Figures



References
-
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279(33):34083–6. - PubMed
-
- Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11(7):672–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

