Exopolyphosphatases PPX1 and PPX2 from Corynebacterium glutamicum
- PMID: 19304823
- PMCID: PMC2681651
- DOI: 10.1128/AEM.02705-08
Exopolyphosphatases PPX1 and PPX2 from Corynebacterium glutamicum
Abstract
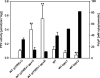
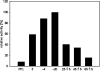
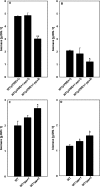
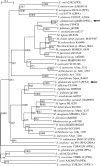
Corynebacterium glutamicum accumulates up to 300 mM of inorganic polyphosphate (PolyP) in the cytosol or in granules. The gene products of cg0488 (ppx1) and cg1115 (ppx2) were shown to be active as exopolyphosphatases (PPX), as overexpression of either gene resulted in higher exopolyphosphatase activities in crude extracts and deletion of either gene with lower activities than those of the wild-type strain. PPX1 and PPX2 from C. glutamicum share only 25% identical amino acids and belong to different protein groups, which are distinct from enterobacterial, archaeal, and yeast exopolyphosphatases. In comparison to that in the wild type, more intracellular PolyP accumulated in the Deltappx1 and Deltappx2 deletion mutations but less when either ppx1 or ppx2 was overexpressed. When C. glutamicum was shifted from phosphate-rich to phosphate-limiting conditions, a growth advantage of the deletion mutants and a growth disadvantage of the overexpression strains compared to the wild type were observed. Growth experiments, exopolyphosphatase activities, and intracellular PolyP concentrations revealed PPX2 as being a major exopolyphosphatase from C. glutamicum. PPX2(His) was purified to homogeneity and shown to be active as a monomer. The enzyme required Mg2+ or Mn2+ cations but was inhibited by millimolar concentrations of Mg2+, Mn2+, and Ca2+. PPX2 from C. glutamicum was active with short-chain polyphosphates, even accepting pyrophosphate, and was inhibited by nucleoside triphosphates.
Figures


Similar articles
-
Functional characterization of exopolyphosphatase/guanosine pentaphosphate phosphohydrolase (PPX/GPPA) of Campylobacter jejuni.Virulence. 2014 May 15;5(4):521-33. doi: 10.4161/viru.28311. Epub 2014 Feb 25. Virulence. 2014. PMID: 24569519 Free PMC article.
-
Ppx1 putative exopolyphosphatase is essential for polyphosphate accumulation in Lacticaseibacillus paracasei.Appl Environ Microbiol. 2024 May 21;90(5):e0229023. doi: 10.1128/aem.02290-23. Epub 2024 Apr 15. Appl Environ Microbiol. 2024. PMID: 38619267 Free PMC article.
-
PPX1 gene overexpression has no influence on polyphosphates in Saccharomyces cerevisiae.Biochemistry (Mosc). 2014 Nov;79(11):1211-5. doi: 10.1134/S000629791411008X. Biochemistry (Mosc). 2014. PMID: 25540006
-
Inorganic polyphosphates and exopolyphosphatases in different cell compartments of Saccharomyces cerevisiae.Biochemistry (Mosc). 2006 Nov;71(11):1171-5. doi: 10.1134/s0006297906110010. Biochemistry (Mosc). 2006. PMID: 17140377 Review.
-
Inorganic polyphosphates in mitochondria.Biochemistry (Mosc). 2010 Jul;75(7):825-31. doi: 10.1134/s0006297910070035. Biochemistry (Mosc). 2010. PMID: 20673205 Review.
Cited by
-
Link between phosphate starvation and glycogen metabolism in Corynebacterium glutamicum, revealed by metabolomics.Appl Environ Microbiol. 2010 Oct;76(20):6910-9. doi: 10.1128/AEM.01375-10. Epub 2010 Aug 27. Appl Environ Microbiol. 2010. PMID: 20802079 Free PMC article.
-
Functional characterization of exopolyphosphatase/guanosine pentaphosphate phosphohydrolase (PPX/GPPA) of Campylobacter jejuni.Virulence. 2014 May 15;5(4):521-33. doi: 10.4161/viru.28311. Epub 2014 Feb 25. Virulence. 2014. PMID: 24569519 Free PMC article.
-
The two PPX-GppA homologues from Mycobacterium tuberculosis have distinct biochemical activities.PLoS One. 2012;7(8):e42561. doi: 10.1371/journal.pone.0042561. Epub 2012 Aug 3. PLoS One. 2012. PMID: 22880033 Free PMC article.
-
Marine Archaeon Methanosarcina acetivorans Enhances Polyphosphate Metabolism Under Persistent Cadmium Stress.Front Microbiol. 2019 Oct 24;10:2432. doi: 10.3389/fmicb.2019.02432. eCollection 2019. Front Microbiol. 2019. PMID: 31708902 Free PMC article.
-
Inorganic Polyphosphates As Storage for and Generator of Metabolic Energy in the Extracellular Matrix.Chem Rev. 2019 Dec 26;119(24):12337-12374. doi: 10.1021/acs.chemrev.9b00460. Epub 2019 Nov 18. Chem Rev. 2019. PMID: 31738523 Free PMC article. Review.
References
-
- Akiyama, M., E. Crooke, and A. Kornberg. 1993. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268:633-639. - PubMed
-
- Ayraud, S., B. Janvier, A. Labigne, C. Ecobichon, C. Burucoa, and J. L. Fauchere. 2005. Polyphosphate kinase: a new colonization factor of Helicobacter pylori. FEMS Microbiol. Lett. 243:45-50. - PubMed
-
- Brown, M. R., and A. Kornberg. 2008. The long and short of it—polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33:284-290. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

