Development of cpn60-based real-time quantitative PCR assays for the detection of 14 Campylobacter species and application to screening of canine fecal samples
- PMID: 19304828
- PMCID: PMC2681665
- DOI: 10.1128/AEM.00101-09
Development of cpn60-based real-time quantitative PCR assays for the detection of 14 Campylobacter species and application to screening of canine fecal samples
Abstract
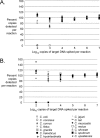
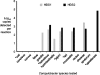
Campylobacter species are important organisms in both human and animal health. The identification of Campylobacter currently requires the growth of organisms from complex samples and biochemical identification. In many cases, the condition of the sample being tested and/or the fastidious nature of many Campylobacter species has limited the detection of campylobacters in a laboratory setting. To address this, we have designed a set of real-time quantitative PCR (qPCR) assays to detect and quantify 14 Campylobacter species, C. coli, C. concisus, C. curvus, C. fetus, C. gracilis, C. helveticus, C. hyointestinalis, C. jejuni, C. lari, C. mucosalis, C. rectus, C. showae, C. sputorum, and C. upsaliensis, directly from DNA extracted from feces. By use of a region of the cpn60 (also known as hsp60 or groEL) gene, which encodes the universally conserved 60-kDa chaperonin, species-specific assays were designed and validated. These assays were then employed to determine the prevalence of Campylobacter species in fecal samples from dogs. Fecal samples were found to contain detectable and quantifiable levels of C. fetus, C. gracilis, C. helveticus, C. jejuni, C. showae, and C. upsaliensis, with the majority of samples containing multiple Campylobacter species. This study represents the first report of C. fetus, C. gracilis, C. mucosalis, and C. showae detection in dogs and implicates dogs as a reservoir for these species. The qPCR assays described offer investigators a new tool to study many Campylobacter species in a culture-independent manner.
Figures
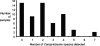
Similar articles
-
Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals.BMC Microbiol. 2010 Mar 10;10:73. doi: 10.1186/1471-2180-10-73. BMC Microbiol. 2010. PMID: 20219122 Free PMC article.
-
A PCR-RFLP assay for the detection and differentiation of Campylobacter jejuni, C. coli, C. fetus, C. hyointestinalis, C. lari, C. helveticus and C. upsaliensis.J Med Microbiol. 2014 May;63(Pt 5):659-666. doi: 10.1099/jmm.0.071498-0. Epub 2014 Feb 25. J Med Microbiol. 2014. PMID: 24568882
-
Use of PCR for direct detection of Campylobacter species in bovine feces.Appl Environ Microbiol. 2003 Jun;69(6):3435-47. doi: 10.1128/AEM.69.6.3435-3447.2003. Appl Environ Microbiol. 2003. PMID: 12788747 Free PMC article.
-
Campylobacteriosis in dogs and cats: a review.N Z Vet J. 2018 Sep;66(5):221-228. doi: 10.1080/00480169.2018.1475268. Epub 2018 May 23. N Z Vet J. 2018. PMID: 29756542 Review.
-
Clinical relevance of infections with zoonotic and human oral species of Campylobacter.J Microbiol. 2016 Jul;54(7):459-67. doi: 10.1007/s12275-016-6254-x. Epub 2016 Jun 28. J Microbiol. 2016. PMID: 27350611 Review.
Cited by
-
Molecular diagnostic tools for detection and differentiation of phytoplasmas based on chaperonin-60 reveal differences in host plant infection patterns.PLoS One. 2014 Dec 31;9(12):e116039. doi: 10.1371/journal.pone.0116039. eCollection 2014. PLoS One. 2014. PMID: 25551224 Free PMC article.
-
Antimicrobial resistant Helicobacter fennelliae isolated from non-diarrheal child stool sample in Battambang, Cambodia.Gut Pathog. 2018 May 30;10:18. doi: 10.1186/s13099-018-0246-9. eCollection 2018. Gut Pathog. 2018. PMID: 29854008 Free PMC article.
-
A Case of Persistent Diarrhea in a Man with the Molecular Detection of Various Campylobacter species and the First Isolation of candidatus Campylobacter infans.Pathogens. 2020 Nov 30;9(12):1003. doi: 10.3390/pathogens9121003. Pathogens. 2020. PMID: 33265947 Free PMC article.
-
Unravelling the reservoirs for colonisation of infants with Campylobacter spp. in rural Ethiopia: protocol for a longitudinal study during a global pandemic and political tensions.BMJ Open. 2022 Oct 5;12(10):e061311. doi: 10.1136/bmjopen-2022-061311. BMJ Open. 2022. PMID: 36198455 Free PMC article.
-
Quantitative Molecular Detection of 19 Major Pathogens in the Interdental Biofilm of Periodontally Healthy Young Adults.Front Microbiol. 2016 Jun 2;7:840. doi: 10.3389/fmicb.2016.00840. eCollection 2016. Front Microbiol. 2016. PMID: 27313576 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- BonDurant, R. H. 2005. Venereal diseases of cattle: natural history, diagnosis, and the role of vaccines in their control. Vet. Clin. N. Am. Food Anim. Pract. 21:383-408. - PubMed
-
- Botteldoorn, N., E. Van Coillie, V. Piessens, G. Rasschaert, L. Debruyne, M. Heyndrickx, L. Herman, and W. Messens. 2008. Quantification of Campylobacter spp. in chicken carcass rinse by real-time PCR. J. Appl. Microbiol. 105:1909-1918. - PubMed
-
- Butzler, J.-P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868-876. - PubMed
-
- Dumonceaux, T. J., J. E. Hill, S. A. Briggs, K. K. Amoako, S. M. Hemmingsen, and A. G. Van Kessel. 2006. Enumeration of specific bacterial populations in complex intestinal communities using quantitative PCR based on the chaperonin-60 target. J. Microbiol. Methods 64:46-62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

