Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis
- PMID: 19304848
- PMCID: PMC2687177
- DOI: 10.1128/JB.01728-08
Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis
Abstract
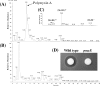
Polymyxin, a long-known peptide antibiotic, has recently been reintroduced in clinical practice because it is sometimes the only available antibiotic for the treatment of multidrug-resistant gram-negative pathogenic bacteria. Lack of information on the biosynthetic genes of polymyxin, however, has limited the study of structure-function relationships and the development of improved polymyxins. During whole genome sequencing of Paenibacillus polymyxa E681, a plant growth-promoting rhizobacterium, we identified a gene cluster encoding polymyxin synthetase. Here, we report the complete sequence of the gene cluster and its function in polymyxin biosynthesis. The gene cluster spanning the 40.6-kb region consists of five open reading frames, designated pmxA, pmxB, pmxC, pmxD, and pmxE. The pmxC and pmxD genes are similar to genes that encode transport proteins, while pmxA, pmxB, and pmxE encode polymyxin synthetases. The insertional disruption of pmxE led to a loss of the ability to produce polymyxin. Introduction of the pmx gene cluster into the amyE locus of the Bacillus subtilis chromosome resulted in the production of polymyxin in the presence of extracellularly added L-2,4-diaminobutyric acid. Taken together, our findings demonstrate that the pmx gene cluster is responsible for polymyxin biosynthesis.
Figures



Similar articles
-
Characterization of the Polymyxin D Synthetase Biosynthetic Cluster and Product Profile of Paenibacillus polymyxa ATCC 10401.J Nat Prod. 2017 May 26;80(5):1264-1274. doi: 10.1021/acs.jnatprod.6b00807. Epub 2017 May 2. J Nat Prod. 2017. PMID: 28463513
-
Biosynthesis of Polymyxins B, E, and P Using Genetically Engineered Polymyxin Synthetases in the Surrogate Host Bacillus subtilis.J Microbiol Biotechnol. 2015 Jul;25(7):1015-25. doi: 10.4014/jmb.1505.05036. J Microbiol Biotechnol. 2015. PMID: 26059516
-
Combinatorial metabolic engineering of Bacillus subtilis for de novo production of polymyxin B.Metab Eng. 2024 May;83:123-136. doi: 10.1016/j.ymben.2024.04.001. Epub 2024 Apr 4. Metab Eng. 2024. PMID: 38582143
-
Chronicle of a Soil Bacterium: Paenibacillus polymyxa E681 as a Tiny Guardian of Plant and Human Health.Front Microbiol. 2019 Mar 15;10:467. doi: 10.3389/fmicb.2019.00467. eCollection 2019. Front Microbiol. 2019. PMID: 30930873 Free PMC article. Review.
-
Regulation of peptide antibiotic production in Bacillus.Mol Microbiol. 1993 Mar;7(5):631-6. doi: 10.1111/j.1365-2958.1993.tb01154.x. Mol Microbiol. 1993. PMID: 7682277 Review.
Cited by
-
Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting Rhizobacterium with broad-spectrum antimicrobial activity.J Bacteriol. 2011 Jan;193(1):311-2. doi: 10.1128/JB.01234-10. Epub 2010 Oct 29. J Bacteriol. 2011. PMID: 21037012 Free PMC article.
-
Fusaricidins, Polymyxins and Volatiles Produced by Paenibacillus polymyxa Strains DSM 32871 and M1.Pathogens. 2021 Nov 15;10(11):1485. doi: 10.3390/pathogens10111485. Pathogens. 2021. PMID: 34832640 Free PMC article.
-
Comparison of Strategies to Overcome Drug Resistance: Learning from Various Kingdoms.Molecules. 2018 Jun 18;23(6):1476. doi: 10.3390/molecules23061476. Molecules. 2018. PMID: 29912169 Free PMC article. Review.
-
Genome Mining and Characterization of Biosynthetic Gene Clusters in Two Cave Strains of Paenibacillus sp.Front Microbiol. 2021 Jan 11;11:612483. doi: 10.3389/fmicb.2020.612483. eCollection 2020. Front Microbiol. 2021. PMID: 33505378 Free PMC article.
-
Acyltransferase that catalyses the condensation of polyketide and peptide moieties of goadvionin hybrid lipopeptides.Nat Chem. 2020 Sep;12(9):869-877. doi: 10.1038/s41557-020-0508-2. Epub 2020 Jul 27. Nat Chem. 2020. PMID: 32719482
References
-
- Ainsworth, G. C., A. M. Brown, and G. Brownlee. 1947. Aerosporin, an antibiotic produced by Bacillus aerosporus Greer. Nature 160263. - PubMed
-
- Alekshun, M. N., and S. B. Levy. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 1281037-1050. - PubMed
-
- Benedict, R. G., and A. F. Langlykke. 1947. Antibiotic activity of Bacillus polymyxa. J. Bacteriol. 5424-25. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

