Automated diagnosis of LC-MS/MS performance
- PMID: 19304874
- PMCID: PMC2677746
- DOI: 10.1093/bioinformatics/btp155
Automated diagnosis of LC-MS/MS performance
Abstract
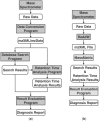
Summary: We report a software scheme for automated diagnosis of liquid chromatography tandem mass spectrometry (LC-MS/MS) system performance. The proposed software scheme provides a robust framework for establishing automated diagnosis of LC-MS/MS system performance for a variety of instruments and experiments. This schematic consists of four main software components: (i) data conversion, (ii) peptide identification, (iii) LC retention time analysis and (iv) system performance evaluation. The implementation of a standard approach for assessing LC-MS/MS system performance enables researchers to apply reliable metrics to assess their workflows performance over different batch experiments. Furthermore, the results from system diagnosis can provide feedback to the workflow to stop batch analysis if system performance falls below prescribed thresholds. A basic implementation of the approach based on the MassMatrix database search and LC retention time analysis programs is presented.
Availability: An open source implementation of the LC-MS/MS system diagnosis software based on the MassMatrix database search program is freely available to non-commercial users and can be downloaded at www.massmatrix.net.
Figures
References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Mischak H, et al. Clinical proteomics: a need to define the field and to begin to set adequate standards. Proteomics Clin. Appl. 2007;1:148–156. - PubMed
-
- Sadygov RG, et al. Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat. Methods. 2004;1:195–202. - PubMed
-
- Shinoda K, et al. Informatics for peptide retention properties in proteomics LC-MS. Proteomics. 2008;8:787–798. - PubMed