Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila
- PMID: 19304924
- PMCID: PMC2673071
- DOI: 10.1261/rna.1364909
Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila
Abstract
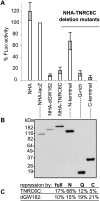
miRNA-mediated repression affects a wide range of biological processes including development and human pathologies. The GW182 protein is a key component of miRNA repression complex, recruited by Argonaute and functioning downstream to repress translation and accelerate mRNA degradation, but little is known about how GW182 proteins act. Using both tethered function and complementation assays, we identify three independent domains of the Drosophila GW182 protein (also termed Gawky) that are sufficient to repress mRNA. Each of these domains also functions independently of poly(A) tails. These results indicate that miRNA-mediated repression is facilitated by multiple domains of GW182.
Figures






References
-
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Bushati N., Cohen S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. - PubMed
-
- Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
