A molecular mechanism of chronological aging in yeast
- PMID: 19305133
- PMCID: PMC2746416
- DOI: 10.4161/cc.8.8.8287
A molecular mechanism of chronological aging in yeast
Abstract
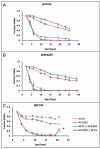
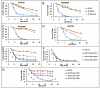
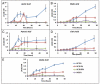
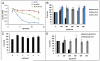
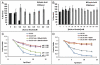
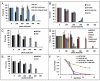
The molecular mechanisms that cause organismal aging are a topic of intense scrutiny and debate. Dietary restriction extends the life span of many organisms, including yeast, and efforts are underway to understand the biochemical and genetic pathways that regulate this life span extension in model organisms. Here we describe the mechanism by which dietary restriction extends yeast chronological life span, defined as the length of time stationary yeast cells remain viable in a quiescent state. We find that aging under standard culture conditions is the result of a cell-extrinsic component that is linked to the pH of the culture medium. We identify acetic acid as a cell-extrinsic mediator of cell death during chronological aging, and demonstrate that dietary restriction, growth in a non-fermentable carbon source, or transferring cells to water increases chronological life span by reducing or eliminating extracellular acetic acid. Other life span extending environmental and genetic interventions, such as growth in high osmolarity media, deletion of SCH9 or RAS2, increase cellular resistance to acetic acid. We conclude that acetic acid induced mortality is the primary mechanism of chronological aging in yeast under standard conditions.
Figures



Comment in
-
Acetic acid effects on aging in budding yeast: are they relevant to aging in higher eukaryotes?Cell Cycle. 2009 Jul 15;8(14):2300-2. doi: 10.4161/cc.8.14.8852. Epub 2009 Jul 27. Cell Cycle. 2009. PMID: 19502784 Free PMC article. No abstract available.
References
-
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. - PubMed
-
- Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–82. - PubMed
-
- Campisi J. Senescent cells, tumor suppression and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–22. - PubMed
-
- Wallenfang MR, Nayak R, DiNardo S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell. 2006;5:297–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
