the need for review and understanding of SELDI/MALDI mass spectroscopy data prior to analysis
- PMID: 19305634
- PMCID: PMC2657646
the need for review and understanding of SELDI/MALDI mass spectroscopy data prior to analysis
Abstract
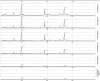
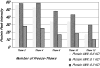
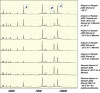
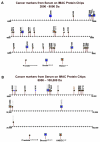
Multiple studies have reported that surface enhanced laser desorption/ionization time of flight mass spectroscopy (SELDI-TOF-MS) is useful in the early detection of disease based on the analysis of bodily fluids. Use of any multiplex mass spectroscopy based approach as in the analysis of bodily fluids to detect disease must be analyzed with great care due to the susceptibility of multiplex and mass spectroscopy methods to biases introduced via experimental design, patient samples, and/or methodology. Specific biases include those related to experimental design, patients, samples, protein chips, chip reader and spectral analysis. Contributions to biases based on patients include demographics (e.g., age, race, ethnicity, sex), homeostasis (e.g., fasting, medications, stress, time of sampling), and site of analysis (hospital, clinic, other). Biases in samples include conditions of sampling (type of sample container, time of processing, time to storage), conditions of storage, (time and temperature of storage), and prior sample manipulation (freeze thaw cycles). Also, there are many potential biases in methodology which can be avoided by careful experimental design including ensuring that cases and controls are analyzed randomly. All the above forms of biases affect any system based on analyzing multiple analytes and especially all mass spectroscopy based methods, not just SELDI-TOF-MS. Also, all current mass spectroscopy systems have relatively low sensitivity compared with immunoassays (e.g., ELISA). There are several problems which may be unique to the SELDI-TOF-MS system marketed by Ciphergen(®). Of these, the most important is a relatively low resolution (±0.2%) of the bundled mass spectrometer which may cause problems with analysis of data. Foremost, this low resolution results in difficulties in determining what constitutes a "peak" if a peak matching approach is used in analysis. Also, once peaks are selected, the peaks may represent multiple proteins. In addition, because peaks may vary slightly in location due to instrumental drift, long term identification of the same peaks may prove to be a challenge. Finally, the Ciphergen(®) system has some "noise" of the baseline which results from the accumulation of charge in the detector system. Thus, we must be very aware of the factors that may affect the use of proteomics in the early detection of disease, in determining aggressive subsets of cancers, in risk assessment and in monitoring the effectiveness of novel therapies.
Keywords: bias; cancer detection; mass spectrometry; serum; specimen processing; specimens.
Figures



References
-
- Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, Wright GL., Jr Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62(13):3609–3614. (#12) - PubMed
-
- Ball G, Mian S, Holding F, Allibone RO, Lowe J, Ali S, Li G, McCardle S, Ellis IO, Creaser C, Rees RC. An integrated approach utilizing artificial neural networks and SELDI mass spectrometry for the classification of human tumoours and rapid identification of potential biomarkers. Bioinformatics. 2002;18:395–404. (#36) - PubMed
-
- Banez LL, Prasanna P, Sun L, Ali A, Zou A, Adam L, McLeod DG, Moul JW, Srivastava S. Diagnostic potential of serum proteomic patterns in prostate cancer. J Urol. 2003;170:442–446. (#14) - PubMed
-
- Carter D, Douglass JF, Cornellison CD, Retter MW, Johnson JC, Bennington AA, Fleming TP, Reed SG, Houghton RL, Diamond DL, Vedvick TS. Purification and characterization of the mammaglobin/lipophilin B complex, a promising diagnostic marker for breast cancer. Biochemistry. 2002;41:6714–6722. (#30) - PubMed
-
- Cazares LH, Adam BL, Ward MD, Nasim S, Schellhammer PF, Semmes OJ, Wright GL., Jr Normal, benign, preneoplastic, and malignant prostate cells have distinct protein expression profiles resolved by surface enhanced laser desorption/ionization mass spectrometry. Clin Cancer Res. 2002;8:2541–2552. (#28) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
