Allosteric theory: taking therapeutic advantage of the malleable nature of GPCRs
- PMID: 19305797
- PMCID: PMC2656818
- DOI: 10.2174/157015907781695973
Allosteric theory: taking therapeutic advantage of the malleable nature of GPCRs
Abstract
The description of the allosteric modification of receptors to affect changes in their function requires a model that considers the effects of the modulator on both agonist affinity and efficacy. A model is presented which describes changes in affinity in terms of the constant alpha (ratio of affinity in the presence vs the absence of modulator) and also the constant xi (ratio of intrinsic efficacy of the agonist in the presence vs absence of modulator). This allows independent effects of both affinity and efficacy and allows the modeling of any change in the dose-response curve to an agonist after treatment with modulator. Examples are given where this type of model can predict effects of modulators that reduce efficacy but actually increase affinity of agonist (i.e. ifenprodil) and also of modulators that block the action of some agonists (the CXCR4 agonist SDF-1alpha by the antagonist AMD3100) but not others for the same receptor (SDF-1alpha peptide fragments RSVM and ASLW).'All models are wrong...but some are useful...'anonymous environmental scientist.
Figures

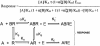


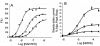


References
-
- Alves ID, Salamon Z, Varga E, Yamamura HI, Tollin G, Hruby VJ. Direct observation of G-protein binding to the human δ-opioid receptor using plasmon-waveguide resonance spectroscopy. J. Biol. Chem. 2003;278:48890–48897. - PubMed
-
- Atchison RE, Gosling J, Monteclaro FS, Franci C, Digilio L, Charo IF, Goldsmith M. Multiple extracellular elements of CCR5 and HIV-1 entry: Dissociation from response to chemokines. Science. 1996;274:1924. - PubMed
-
- Black JW, Leff P. Operational models of pharmacological agonist. Proc. R. Soc. Lond. (Biol.) 1983;220:141–162. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
