Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome
- PMID: 19306292
- PMCID: PMC2731485
- DOI: 10.1002/mc.20499
Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome
Abstract
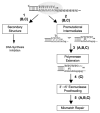
Microsatellite sequences are ubiquitous in the human genome and are important regulators of genome function. Here, we examine the mutational mechanisms governing the stability of highly abundant mono-, di-, and tetranucleotide microsatellites. Microsatellite mutation rate estimates from pedigree analyses and experimental models range from a low of approximately 10(-6) to a high of approximately 10(-2) mutations per locus per generation. The vast majority of observed mutational variation can be attributed to features intrinsic to the allele itself, including motif size, length, and sequence composition. A greater than linear relationship between motif length and mutagenesis has been observed in several model systems. Motif sequence differences contribute up to 10-fold to the variation observed in human cell mutation rates. The major mechanism of microsatellite mutagenesis is strand slippage during DNA synthesis. DNA polymerases produce errors within microsatellites at a frequency that is 10- to 100-fold higher than the frequency of frameshifts in coding sequences. Motif sequence significantly affects both polymerase error rate and specificity, resulting in strand biases within complementary microsatellites. Importantly, polymerase errors within microsatellites include base substitutions, deletions, and complex mutations, all of which produced interrupted alleles from pure microsatellites. Postreplication mismatch repair efficiency is affected by microsatellite motif size and sequence, also contributing to the observed variation in microsatellite mutagenesis. Inhibition of DNA synthesis within common microsatellites is highly sequence-dependent, and is positively correlated with the production of errors. DNA secondary structure within common microsatellites can account for some DNA polymerase pause sites, and may be an important factor influencing mutational specificity.
(c) 2009 Wiley-Liss, Inc.
Figures


References
-
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11:2453–2465. - PubMed
-
- Katti MV, Ranjekar PK, Gupta VS. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Molecular Biology and Evolution. 2001;18:1161–1167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

