Gene silencing below the immune radar
- PMID: 19306498
- PMCID: PMC2648672
- DOI: 10.1172/jci38475
Gene silencing below the immune radar
Abstract
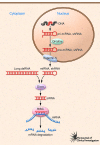
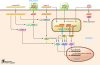
In vertebrates, the detection of viral nucleic acids is the first step toward innate and subsequent adaptive antiviral immune responses. A sophisticated,protein receptor-based sensor system has evolved to recognize viral nucleic acids and to trigger a variety of antiviral defense mechanisms. The more we learn about this elaborate sensor system, the more it becomes evident how difficult it is to introduce exogenous nucleic acids such as siRNA into cells without triggering antiviral immunoreceptors. In this issue of the JCI, Judge and colleagues provide evidence that siRNA can be designed and delivered in a way that allows specific and successful silencing of target genes in tumor cells in vivo, leading to tumor cell death and prolonged survival of tumor-bearing mice in the absence of immune activation. This study represents a major technological advance, setting new standards for well-controlled siRNA applications in vivo, and has the potential to guide clinical development toward siRNA therapeutics with well-defined and selective gene-silencing activities.
Figures
Comment on
-
Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice.J Clin Invest. 2009 Mar;119(3):661-73. doi: 10.1172/JCI37515. Epub 2009 Feb 23. J Clin Invest. 2009. PMID: 19229107 Free PMC article.

