Engineered human antibody constant domains with increased stability
- PMID: 19307178
- PMCID: PMC2682868
- DOI: 10.1074/jbc.M900769200
Engineered human antibody constant domains with increased stability
Abstract
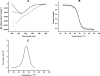
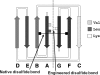
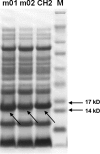
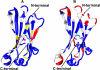
The immunoglobulin (Ig) constant CH2 domain is critical for antibody effector functions. Isolated CH2 domains are promising as scaffolds for construction of libraries containing diverse binders that could also confer some effector functions. However, previous work has shown that an isolated murine CH2 domain is relatively unstable to thermally induced unfolding. To explore unfolding mechanisms of isolated human CH2 and increase its stability gamma1 CH2 was cloned and a panel of cysteine mutants was constructed. Human gamma1 CH2 unfolded at a higher temperature (T(m) = 54.1 degrees C, as measured by circular dichroism) than that previously reported for a mouse CH2 (41 degrees C). One mutant (m01) was remarkably stable (T(m) = 73.8 degrees C). Similar results were obtained by differential scanning calorimetry. This mutant was also significantly more stable than the wild-type CH2 against urea induced unfolding (50% unfolding at urea concentration of 6.8 m versus 4.2 m). The m01 was highly soluble and monomeric. The existence of the second disulfide bond in m01 and its correct position were demonstrated by mass spectrometry and nuclear magnetic resonance spectroscopy, respectively. The loops were on average more flexible than the framework in both CH2 and m01, and the overall secondary structure was not affected by the additional disulfide bond. These data suggest that a human CH2 domain is relatively stable to unfolding at physiological temperature, and that both CH2 and the highly stable mutant m01 are promising new scaffolds for the development of therapeutics against human diseases.
Figures


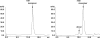

References
-
- Kolmar, H., and Skerra, A. (2008) FEBS J. 275 2667. - PubMed
-
- Skerra, A. (2007) Curr. Opin. Biotechnol. 18 295–304 - PubMed
-
- Nygren, P. A., and Skerra, A. (2004) J. Immunol. Methods 290 3–28 - PubMed
-
- Binz, H. K., Amstutz, P., and Pluckthun, A. (2005) Nat. Biotechnol. 23 1257–1268 - PubMed
-
- Hey, T., Fiedler, E., Rudolph, R., and Fiedler, M. (2005) Trends Biotechnol. 23 514–522 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

