Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia
- PMID: 19307190
- PMCID: PMC2685307
- DOI: 10.1093/aob/mcp058
Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia
Abstract
Background and methods: Polyploidy results in genetic turmoil, much of which is associated with new phenotypes that result in speciation. Five independent lines of synthetic allotetraploid N. x obtusiata (N x o) were created from crosses between the diploid N. attenuata (Na) (male) and N. obtusifolia (No) (female) and the autotetraploids of Na (NaT) and No (NoT) were synthesized. Their genetic, genomic and phenotypic changes were then compared with those of the parental diploid species (Na and No) as well as to the natural allotetraploids, N. quadrivalvis (Nq) and N. clevelandii (Nc), which formed 1 million years ago from crosses between ancient Na and No.
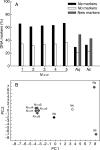
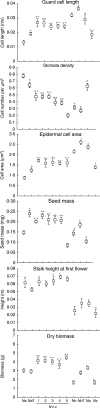
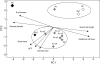
Key results: DNA fingerprinting profiles (by UP-PCR) revealed that the five N x o lines shared similar but not identical profiles. Both synthetic and natural polyploidy showed a dosage effect on genome size (as measured in seeds); however, only Nq was associated with a genome upsizing. Phenotypic analysis revealed that at the cellular level, N x o lines had phenotypes intermediate of the parental phenotypes. Both allo- and autotetraploidization had a dosage effect on seed and dry biomass (except for NaT), but not on stalk height at first flower. Nc showed paternal (Na) cellular phenotypes but inherited maternal (No) biomass and seed mass, whereas Nq showed maternal (No) cellular phenotypes but inherited paternal (Na) biomass and seed mass patterns. Principal component analysis grouped Nq with N x o lines, due to similar seed mass, stalk height and genome size. These traits separated Nc, No and Na from Nq and N x o lines, whereas biomass distinguished Na from N x o and Nq lines, and NaT clustered closer to Nq and N x o lines than to Na.
Conclusions: Both allo- and autotetraploidy induce considerable morphological, genetic and genomic changes, many of which are retained by at least one of the natural polyploids. It is proposed that both natural and synthetic polyploids are well suited for studying the evolution of adaptive responses.
Figures




References
-
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Baldwin IT, Staszakkozinski L, Davidson R. Up in smoke. I. Smoke-derived germination cues for post-fire annual, Nicotiana attenuata torr. ex. Watson. Journal of Chemical Ecology. 1994;20:2345–2371. - PubMed
-
- Beck SL, Dunlop RW, Fossey A. Stomatal length and frequency as a measure of ploidy level in black wattle, Acacia mearnsii (de Wild) Botanical Journal of the Linnean Society. 2003;141:177–181.
-
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: 583 new estimates. Annals of Botany. 1997;80:169–196.
-
- Bulat SA, Lubeck M, Alekhina IA, Jensen DF, Knudsen IMB, Lubeck PS. Identification of a universally primed-PCR-derived sequence-characterized amplified region marker for an antagonistic strain of Clonostachys rosea and development of a strain-specific PCR detection assay. Applied and Environmental Microbiology. 2000;66:4758–4763. - PMC - PubMed

