The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm
- PMID: 19307375
- PMCID: PMC2687228
- DOI: 10.1128/AAC.00043-09
The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm
Abstract
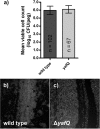
Escherichia coli is refractory to elevated doses of antibiotics when it is growing in a biofilm, and this is potentially due to high numbers of multidrug-tolerant persister cells in the surface-adherent population. Previously, the chromosomal toxin-antitoxin loci hipBA and relBE have been linked to the frequency at which persister cells occur in E. coli populations. In the present study, we focused on the dinJ-yafQ-encoded toxin-antitoxin system and hypothesized that deletion of the toxin gene yafQ might influence cell survival in antibiotic-exposed biofilms. By using confocal laser scanning microscopy and viable cell counting, it was determined that a Delta yafQ mutant produced biofilms with a structure and a cell density equivalent to those of the parental strain. In-depth susceptibility testing identified that relative to wild-type E. coli, the Delta yafQ strain had up to a approximately 2,400-fold decrease in cell survival after the biofilms were exposed to bactericidal concentrations of cefazolin or tobramycin. Corresponding to these data, controlled overexpression of yafQ from a high-copy-number plasmid resulted in up to a approximately 10,000-fold increase in the number of biofilm cells surviving exposure to these bactericidal drugs. In contrast, neither the inactivation nor the overexpression of yafQ affected the tolerance of biofilms to doxycycline or rifampin (rifampicin). Furthermore, deletion of yafQ did not affect the tolerance of stationary-phase planktonic cells to any of the antibacterials tested. These results suggest that yafQ mediates the tolerance of E. coli biofilms to multiple but specific antibiotics; moreover, our data imply that this cellular pathway for persistence is likely different from that of multidrug-tolerant cells in stationary-phase planktonic cell cultures.
Figures





References
-
- Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. - PubMed
-
- Bigger, J. W. 1944. Treatment of staphylococcal infections with penicillin. Lancet ii:497-500.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

