Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing
- PMID: 19307605
- PMCID: PMC2674820
- DOI: 10.1534/genetics.109.101998
Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing
Abstract
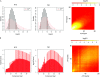
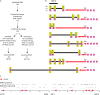
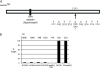
Next-generation methods for rapid whole-genome sequencing enable the identification of single-base-pair mutations in Drosophila by comparing a chromosome bearing a new mutation to the unmutagenized sequence. To validate this approach, we sought to identify the molecular lesion responsible for a recessive EMS-induced mutation affecting egg shell morphology by using Illumina next-generation sequencing. After obtaining sufficient sequence from larvae that were homozygous for either wild-type or mutant chromosomes, we obtained high-quality reads for base pairs composing approximately 70% of the third chromosome of both DNA samples. We verified 103 single-base-pair changes between the two chromosomes. Nine changes were nonsynonymous mutations and two were nonsense mutations. One nonsense mutation was in a gene, encore, whose mutations produce an egg shell phenotype also observed in progeny of homozygous mutant mothers. Complementation analysis revealed that the chromosome carried a new functional allele of encore, demonstrating that one round of next-generation sequencing can identify the causative lesion for a phenotype of interest. This new method of whole-genome sequencing represents great promise for mutant mapping in flies, potentially replacing conventional methods.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

