The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy
- PMID: 19307726
- PMCID: PMC2662564
- DOI: 10.1172/JCI37630
The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy
Abstract
gamma-Retroviral vectors (gammaRVs), which are commonly used in gene therapy, can trigger oncogenesis by insertional mutagenesis. Here, we have dissected the contribution of vector design and viral integration site selection (ISS) to oncogenesis using an in vivo genotoxicity assay based on transplantation of vector-transduced tumor-prone mouse hematopoietic stem/progenitor cells. By swapping genetic elements between gammaRV and lentiviral vectors (LVs), we have demonstrated that transcriptionally active long terminal repeats (LTRs) are major determinants of genotoxicity even when reconstituted in LVs and that self-inactivating (SIN) LTRs enhance the safety of gammaRVs. By comparing the genotoxicity of vectors with matched active LTRs, we were able to determine that substantially greater LV integration loads are required to approach the same oncogenic risk as gammaRVs. This difference in facilitating oncogenesis is likely to be explained by the observed preferential targeting of cancer genes by gammaRVs. This integration-site bias was intrinsic to gammaRVs, as it was also observed for SIN gammaRVs that lacked genotoxicity in our model. Our findings strongly support the use of SIN viral vector platforms and show that ISS can substantially modulate genotoxicity.
Figures
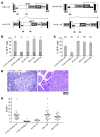
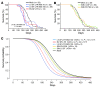
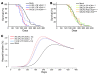

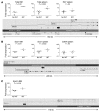

Comment in
-
Preventing and exploiting the oncogenic potential of integrating gene vectors.J Clin Invest. 2009 Apr;119(4):755-8. doi: 10.1172/jci38831. J Clin Invest. 2009. PMID: 19348042 Free PMC article.
Similar articles
-
Foamy Virus Vector Carries a Strong Insulator in Its Long Terminal Repeat Which Reduces Its Genotoxic Potential.J Virol. 2017 Dec 14;92(1):e01639-17. doi: 10.1128/JVI.01639-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29046446 Free PMC article.
-
Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity.Mol Ther. 2012 May;20(5):1022-32. doi: 10.1038/mt.2011.309. Epub 2012 Feb 14. Mol Ther. 2012. PMID: 22334016 Free PMC article.
-
Comparative clonal analysis of reconstitution kinetics after transplantation of hematopoietic stem cells gene marked with a lentiviral SIN or a γ-retroviral LTR vector.Exp Hematol. 2013 Jan;41(1):28-38.e3. doi: 10.1016/j.exphem.2012.09.003. Epub 2012 Sep 16. Exp Hematol. 2013. PMID: 22989760
-
Current landscape of vector safety and genotoxicity after hematopoietic stem or immune cell gene therapy.Leukemia. 2025 Jun;39(6):1325-1333. doi: 10.1038/s41375-025-02585-8. Epub 2025 Apr 8. Leukemia. 2025. PMID: 40200078 Free PMC article. Review.
-
Viral Vectors: The Road to Reducing Genotoxicity.Toxicol Sci. 2017 Feb;155(2):315-325. doi: 10.1093/toxsci/kfw220. Epub 2016 Nov 1. Toxicol Sci. 2017. PMID: 27803388 Review.
Cited by
-
A nonintegrative lentiviral vector-based vaccine provides long-term sterile protection against malaria.PLoS One. 2012;7(11):e48644. doi: 10.1371/journal.pone.0048644. Epub 2012 Nov 2. PLoS One. 2012. PMID: 23133649 Free PMC article.
-
Foamy viral vector integration sites in SCID-repopulating cells after MGMTP140K-mediated in vivo selection.Gene Ther. 2015 Jul;22(7):591-5. doi: 10.1038/gt.2015.20. Epub 2015 Mar 19. Gene Ther. 2015. PMID: 25786870 Free PMC article.
-
Correcting inborn errors of immunity: From viral mediated gene addition to gene editing.Semin Immunol. 2023 Mar;66:101731. doi: 10.1016/j.smim.2023.101731. Epub 2023 Feb 28. Semin Immunol. 2023. PMID: 36863140 Free PMC article. Review.
-
Chicken HS4 insulators have minimal barrier function among progeny of human hematopoietic cells transduced with an HIV1-based lentiviral vector.Mol Ther. 2011 Jan;19(1):133-9. doi: 10.1038/mt.2010.218. Epub 2010 Oct 12. Mol Ther. 2011. PMID: 20940706 Free PMC article.
-
A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells.Blood. 2010 Aug 12;116(6):900-8. doi: 10.1182/blood-2009-10-250209. Epub 2010 May 10. Blood. 2010. PMID: 20457870 Free PMC article.
References
-
- Will E., et al. Importance of murine study design for testing toxicity of retroviral vectors in support of phase I trials. Mol. Ther. 2007;15:782–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

