Kallikrein genes are associated with lupus and glomerular basement membrane-specific antibody-induced nephritis in mice and humans
- PMID: 19307730
- PMCID: PMC2662554
- DOI: 10.1172/JCI36728
Kallikrein genes are associated with lupus and glomerular basement membrane-specific antibody-induced nephritis in mice and humans
Abstract
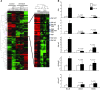
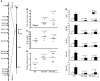
Immune-mediated nephritis contributes to disease in systemic lupus erythematosus, Goodpasture syndrome (caused by antibodies specific for glomerular basement membrane [anti-GBM antibodies]), and spontaneous lupus nephritis. Inbred mouse strains differ in susceptibility to anti-GBM antibody-induced and spontaneous lupus nephritis. This study sought to clarify the genetic and molecular factors that maybe responsible for enhanced immune-mediated renal disease in these models. When the kidneys of 3 mouse strains sensitive to anti-GBM antibody-induced nephritis were compared with those of 2 control strains using microarray analysis, one-fifth of the underexpressed genes belonged to the kallikrein gene family,which encodes serine esterases. Mouse strains that upregulated renal and urinary kallikreins exhibited less evidence of disease. Antagonizing the kallikrein pathway augmented disease, while agonists dampened the severity of anti-GBM antibody-induced nephritis. In addition, nephritis-sensitive mouse strains had kallikrein haplotypes that were distinct from those of control strains, including several regulatory polymorphisms,some of which were associated with functional consequences. Indeed, increased susceptibility to anti-GBM antibody-induced nephritis and spontaneous lupus nephritis was achieved by breeding mice with a genetic interval harboring the kallikrein genes onto a disease-resistant background. Finally, both human SLE and spontaneous lupus nephritis were found to be associated with kallikrein genes, particularly KLK1 and the KLK3 promoter, when DNA SNPs from independent cohorts of SLE patients and controls were compared. Collectively, these studies suggest that kallikreins are protective disease-associated genes in anti-GBM antibody-induced nephritis and lupus.
Figures
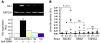



Comment in
-
Kallikreins and lupus nephritis.J Clin Invest. 2009 Apr;119(4):768-71. doi: 10.1172/jci38786. J Clin Invest. 2009. PMID: 19348047 Free PMC article.
References
-
- Foster M.H., Kelley V.R. Lupus nephritis: update on pathogenesis and disease mechanisms. . Semin. Nephrol. 1999;19:173–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

