MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells
- PMID: 19309508
- PMCID: PMC2669448
- DOI: 10.1186/1479-5876-7-20
MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells
Abstract
Background: The unique features of human embryonic stem (hES) cells make them the best candidate resource for both cell replacement therapy and development research. However, the molecular mechanisms responsible for the simultaneous maintenance of their self-renewal properties and undifferentiated state remain unclear. Non-coding microRNAs (miRNA) which regulate mRNA cleavage and inhibit encoded protein translation exhibit temporal or tissue-specific expression patterns and they play an important role in development timing.
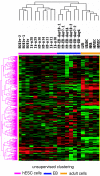
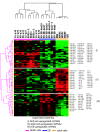
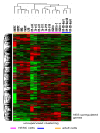
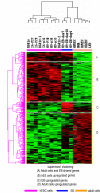
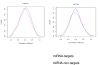
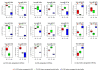
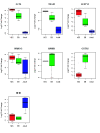
Results: In this study, we analyzed miRNA and gene expression profiles among samples from 3 hES cell lines (H9, I6 and BG01v), differentiated embryoid bodies (EB) derived from H9 cells at different time points, and 5 adult cell types including Human Microvascular Endothelial Cells (HMVEC), Human Umbilical Vein Endothelial Cells (HUVEC), Umbilical Artery Smooth Muscle Cells (UASMC), Normal Human Astrocytes (NHA), and Lung Fibroblasts (LFB). This analysis rendered 104 miRNAs and 776 genes differentially expressed among the three cell types. Selected differentially expressed miRNAs and genes were further validated and confirmed by quantitative real-time-PCR (qRT-PCR). Especially, members of the miR-302 cluster on chromosome 4 and miR-520 cluster on chromosome 19 were highly expressed in undifferentiated hES cells. MiRNAs in these two clusters displayed similar expression levels. The members of these two clusters share a consensus 7-mer seed sequence and their targeted genes had overlapping functions. Among the targeted genes, genes with chromatin structure modification function are enriched suggesting a role in the maintenance of chromatin structure. We also found that the expression level of members of the two clusters, miR-520b and miR-302c, were negatively correlated with their targeted genes based on gene expression analysis
Conclusion: We identified the expression patterns of miRNAs and gene transcripts in the undifferentiation of human embryonic stem cells; among the miRNAs that are highly expressed in undifferentiated embryonic stem cells, the miR-520 cluster may be closely involved in hES cell function and its relevance to chromatin structure warrants further study.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

