Mutational analysis of Mycobacterium UvrD1 identifies functional groups required for ATP hydrolysis, DNA unwinding, and chemomechanical coupling
- PMID: 19317511
- PMCID: PMC2761027
- DOI: 10.1021/bi900103d
Mutational analysis of Mycobacterium UvrD1 identifies functional groups required for ATP hydrolysis, DNA unwinding, and chemomechanical coupling
Abstract
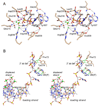
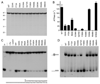
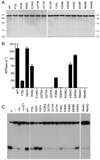
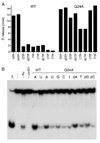
Mycobacterial UvrD1 is a DNA-dependent ATPase and a Ku-dependent 3' to 5' DNA helicase. The UvrD1 motor domain resembles that of the prototypal superfamily I helicases UvrD and PcrA. Here we performed a mutational analysis of UvrD1 guided by the crystal structure of a DNA-bound Escherichia coli UvrD-ADP-MgF(3) transition state mimetic. Alanine scanning and conservative substitutions identified amino acids essential for both ATP hydrolysis and duplex unwinding, including those implicated in phosphohydrolase chemistry via transition state stabilization (Arg308, Arg648, Gln275), divalent cation coordination (Glu236), or activation of the nucleophilic water (Glu236, Gln275). Other residues important for ATPase/helicase activity include Phe280 and Phe72, which interact with the DNA 3' single strand tail. ATP hydrolysis was uncoupled from duplex unwinding by mutations at Glu609 (in helicase motif V), which contacts the ATP ribose sugar. Introducing alanine in lieu of the adenine-binding "Q motif" glutamine (Gln24) relaxed the substrate specificity in NTP hydrolysis, e.g., eliciting a gain of function as a UTPase/TTPase, although the Q24A mutant still relied on ATP/dATP for duplex unwinding. Our studies highlight the role of the Q motif as a substrate filter and the contributions of adenosine-binding residues as couplers of NTP hydrolysis to motor activity. The Ku-binding function of UvrD1 lies within its C-terminal 270 amino acid segment. Here we found that deleting the 90 amino acid C-terminal domain, which is structurally uncharacterized, diminished DNA unwinding, without affecting ATP hydrolysis or binding to the DNA helicase substrate, apparently by affecting the strength of the UvrD1-Ku interaction.
Figures


References
-
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. - PubMed
-
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. - PubMed
-
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structure of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;87:75–84. - PubMed
-
- Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

