Inactivation of Mycobacterium tuberculosis mannosyltransferase pimB reduces the cell wall lipoarabinomannan and lipomannan content and increases the rate of bacterial-induced human macrophage cell death
- PMID: 19318518
- PMCID: PMC2688391
- DOI: 10.1093/glycob/cwp042
Inactivation of Mycobacterium tuberculosis mannosyltransferase pimB reduces the cell wall lipoarabinomannan and lipomannan content and increases the rate of bacterial-induced human macrophage cell death
Abstract
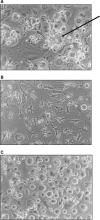
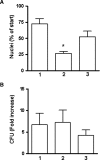
The Mycobacterium tuberculosis (M.tb) cell wall contains an important group of structurally related mannosylated lipoglycans called phosphatidyl-myo-inositol mannosides (PIMs), lipomannan (LM), and mannose-capped lipoarabinomannan (ManLAM), where the terminal alpha-[1-->2] mannosyl structures on higher order PIMs and ManLAM have been shown to engage C-type lectins such as the macrophage mannose receptor directing M.tb phagosome maturation arrest. An important gene described in the biosynthesis of these molecules is the mannosyltransferase pimB (Rv0557). Here, we disrupted pimB in a virulent strain of M.tb. We demonstrate that the inactivation of pimB in M.tb does not abolish the production of any of its cell wall mannosylated lipoglycans; however, it results in a quantitative decrease in the ManLAM and LM content without affecting higher order PIMs. This finding indicates gene redundancy or the possibility of an alternative biosynthetic pathway that may compensate for the PimB deficiency. Furthermore, infection of human macrophages by the pimB mutant leads to an alteration in macrophage phenotype concomitant with a significant increase in the rate of macrophage death.
Figures





Similar articles
-
The Phosphatidyl-myo-Inositol Dimannoside Acyltransferase PatA Is Essential for Mycobacterium tuberculosis Growth In Vitro and In Vivo.J Bacteriol. 2021 Mar 8;203(7):e00439-20. doi: 10.1128/JB.00439-20. Print 2021 Mar 8. J Bacteriol. 2021. PMID: 33468587 Free PMC article.
-
Inactivation of Corynebacterium glutamicum NCgl0452 and the role of MgtA in the biosynthesis of a novel mannosylated glycolipid involved in lipomannan biosynthesis.J Biol Chem. 2007 Feb 16;282(7):4561-4572. doi: 10.1074/jbc.M608695200. Epub 2006 Dec 19. J Biol Chem. 2007. PMID: 17179146
-
Fine discrimination in the recognition of individual species of phosphatidyl-myo-inositol mannosides from Mycobacterium tuberculosis by C-type lectin pattern recognition receptors.J Immunol. 2006 Aug 1;177(3):1805-16. doi: 10.4049/jimmunol.177.3.1805. J Immunol. 2006. PMID: 16849491
-
Lipoarabinomannan, and its related glycolipids, induce divergent and opposing immune responses to Mycobacterium tuberculosis depending on structural diversity and experimental variations.Tuberculosis (Edinb). 2016 Jan;96:120-30. doi: 10.1016/j.tube.2015.09.005. Epub 2015 Oct 28. Tuberculosis (Edinb). 2016. PMID: 26586646 Review.
-
Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response.Mol Microbiol. 2004 Jul;53(2):391-403. doi: 10.1111/j.1365-2958.2004.04183.x. Mol Microbiol. 2004. PMID: 15228522 Review.
Cited by
-
The Phosphatidyl-myo-Inositol Dimannoside Acyltransferase PatA Is Essential for Mycobacterium tuberculosis Growth In Vitro and In Vivo.J Bacteriol. 2021 Mar 8;203(7):e00439-20. doi: 10.1128/JB.00439-20. Print 2021 Mar 8. J Bacteriol. 2021. PMID: 33468587 Free PMC article.
-
Mannose-capped lipoarabinomannan in Mycobacterium tuberculosis pathogenesis.Pathog Dis. 2018 Jun 1;76(4):fty026. doi: 10.1093/femspd/fty026. Pathog Dis. 2018. PMID: 29722821 Free PMC article. Review.
-
Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction.FEMS Microbiol Rev. 2011 Nov;35(6):1126-57. doi: 10.1111/j.1574-6976.2011.00276.x. Epub 2011 May 31. FEMS Microbiol Rev. 2011. PMID: 21521247 Free PMC article. Review.
-
Diagnosis of paediatric tuberculosis by optically detecting two virulence factors on extracellular vesicles in blood samples.Nat Biomed Eng. 2022 Aug;6(8):979-991. doi: 10.1038/s41551-022-00922-1. Epub 2022 Aug 19. Nat Biomed Eng. 2022. PMID: 35986185 Free PMC article.
-
Comparative transcriptional study of the putative mannose donor biosynthesis genes in virulent Mycobacterium tuberculosis and attenuated Mycobacterium bovis BCG strains.Infect Immun. 2011 Nov;79(11):4668-73. doi: 10.1128/IAI.05635-11. Epub 2011 Sep 6. Infect Immun. 2011. PMID: 21896775 Free PMC article.
References
-
- Appelmelk BJ, den DJ, Driessen NN, Ummels R, Pak M, Nigou J, Larrouy-Maumus G, Gurcha SS, Movahedzadeh F, Geurtsen J, et al. The mannose cap of mycobacterial lipoarabinomannan does not dominate the Mycobacterium–host interaction. Cell Microbiol. 2008;10:930–944. - PubMed
-
- Barry CE III. Interpreting cell wall “virulence factors” of Mycobacterium tuberculosis. Trends Microbiol. 2001;9:237–241. - PubMed
-
- Baulard AR, Gurcha SS, Engohang-Ndong J, Gouffi K, Locht C, Besra GS. In vivo interaction between the polyprenol phosphate mannose synthase Ppm1 and the integral membrane protein Ppm2 from Mycobacterium smegmatis revealed by a bacterial two-hybrid system. J Biol Chem. 2003;278:2242–2248. - PubMed
-
- Berg S, Kaur D, Jackson M, Brennan PJ. The glycosyltransferases of Mycobacterium tuberculosis: Roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology. 2007;17:35–56R. - PubMed

