Molecular evolution of the junctophilin gene family
- PMID: 19318539
- PMCID: PMC2685503
- DOI: 10.1152/physiolgenomics.00017.2009
Molecular evolution of the junctophilin gene family
Abstract
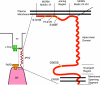
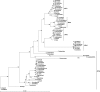
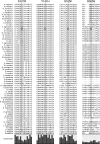
Junctophilins (JPHs) are members of a junctional membrane complex protein family important for the physical approximation of plasmalemmal and sarcoplasmic/endoplasmic reticulum membranes. As such, JPHs facilitate signal transduction in excitable cells between plasmalemmal voltage-gated calcium channels and intracellular calcium release channels. To determine the molecular evolution of the JPH gene family, we performed a phylogenetic analysis of over 60 JPH genes from over 40 species and compared conservation across species and different isoforms. We found that JPHs are evolutionary highly conserved, in particular the membrane occupation and recognition nexus motifs found in all species. Our data suggest that an ancestral form of JPH arose at the latest in a common metazoan ancestor and that in vertebrates four isoforms arose, probably following two rounds of whole genome duplications. By combining multiple prediction techniques with sequence alignments, we also postulate the presence of new important functional regions and candidate sites for posttranslational modifications. The increasing number of available sequences yields significant insight into the molecular evolution of JPHs. Our analysis is consistent with the emerging concept that JPHs serve dual important functions in excitable cells: structural assembly of junctional membrane complexes and regulation of intracellular calcium signaling pathways.
Figures

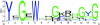

References
-
- Altschul SF, Gish W. Local alignment statistics. Methods Enzymol 266: 460–480, 1996. - PubMed
-
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55: 539–552, 2006. - PubMed
-
- Bers DM Macromolecular complexes regulating cardiac ryanodine receptor function. J Mol Cell Cardiol 37: 417–429, 2004. - PubMed
-
- Brittsan AG, Kranias EG. Phospholamban and cardiac contractile function. J Mol Cell Cardiol 32: 2131–2139, 2000. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

