Correlation of microRNA levels during hypoxia with predicted target mRNAs through genome-wide microarray analysis
- PMID: 19320992
- PMCID: PMC2667434
- DOI: 10.1186/1755-8794-2-15
Correlation of microRNA levels during hypoxia with predicted target mRNAs through genome-wide microarray analysis
Abstract
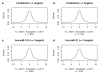
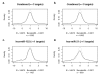
Background: Low levels of oxygen in tissues, seen in situations such as chronic lung disease, necrotic tumors, and high altitude exposures, initiate a signaling pathway that results in active transcription of genes possessing a hypoxia response element (HRE). The aim of this study was to investigate whether a change in miRNA expression following hypoxia could account for changes in the cellular transcriptome based on currently available miRNA target prediction tools.
Methods: To identify changes induced by hypoxia, we conducted mRNA- and miRNA-array-based experiments in HT29 cells, and performed comparative analysis of the resulting data sets based on multiple target prediction algorithms. To date, few studies have investigated an environmental perturbation for effects on genome-wide miRNA levels, or their consequent influence on mRNA output.
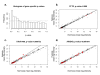
Results: Comparison of miRNAs with predicted mRNA targets indicated a lower level of concordance than expected. We did, however, find preliminary evidence of combinatorial regulation of mRNA expression by miRNA.
Conclusion: Target prediction programs and expression profiling techniques do not yet adequately represent the complexity of miRNA-mediated gene repression, and new methods may be required to better elucidate these pathways. Our data suggest the physiologic impact of miRNAs on cellular transcription results from a multifaceted network of miRNA and mRNA relationships, working together in an interconnected system and in context of hundreds of RNA species. The methods described here for comparative analysis of cellular miRNA and mRNA will be useful for understanding genome wide regulatory responsiveness and refining miRNA predictive algorithms.
Figures








Similar articles
-
Regulatory Network Analysis in Estradiol-Treated Human Endothelial Cells.Int J Mol Sci. 2021 Jul 30;22(15):8193. doi: 10.3390/ijms22158193. Int J Mol Sci. 2021. PMID: 34360960 Free PMC article.
-
Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC.J Transl Med. 2019 Jan 3;17(1):7. doi: 10.1186/s12967-018-1761-7. J Transl Med. 2019. PMID: 30602391 Free PMC article.
-
Inter- and intra-combinatorial regulation by transcription factors and microRNAs.BMC Genomics. 2007 Oct 30;8:396. doi: 10.1186/1471-2164-8-396. BMC Genomics. 2007. PMID: 17971223 Free PMC article.
-
Quantum Language of MicroRNA: Application for New Cancer Therapeutic Targets.Methods Mol Biol. 2018;1733:145-157. doi: 10.1007/978-1-4939-7601-0_12. Methods Mol Biol. 2018. PMID: 29435930
-
miRNAs associated with immune response in teleost fish.Dev Comp Immunol. 2017 Oct;75:77-85. doi: 10.1016/j.dci.2017.02.023. Epub 2017 Feb 28. Dev Comp Immunol. 2017. PMID: 28254620 Review.
Cited by
-
MicroRNA profiling in ischemic injury of the gracilis muscle in rats.BMC Musculoskelet Disord. 2010 Jun 17;11:123. doi: 10.1186/1471-2474-11-123. BMC Musculoskelet Disord. 2010. PMID: 20553627 Free PMC article.
-
MicroRNA response to hypoxic stress in soft tissue sarcoma cells: microRNA mediated regulation of HIF3α.BMC Cancer. 2014 Jun 13;14:429. doi: 10.1186/1471-2407-14-429. BMC Cancer. 2014. PMID: 24927770 Free PMC article.
-
miR-200b downregulates CFTR during hypoxia in human lung epithelial cells.Cell Mol Biol Lett. 2017 Nov 15;22:23. doi: 10.1186/s11658-017-0054-0. eCollection 2017. Cell Mol Biol Lett. 2017. PMID: 29167681 Free PMC article.
-
Opposing roles for HIF-1α and HIF-2α in the regulation of angiogenesis by mononuclear phagocytes.Blood. 2011 Jan 6;117(1):323-32. doi: 10.1182/blood-2010-01-261792. Epub 2010 Oct 15. Blood. 2011. PMID: 20952691 Free PMC article.
-
Cell type-specific response of colon cancer tumor cell lines to oncolytic HSV-1 virotherapy in hypoxia.Cancer Cell Int. 2022 Apr 27;22(1):164. doi: 10.1186/s12935-022-02564-4. Cancer Cell Int. 2022. PMID: 35477503 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

