In vivo stable isotope labeling of fruit flies reveals post-transcriptional regulation in the maternal-to-zygotic transition
- PMID: 19321433
- PMCID: PMC2709187
- DOI: 10.1074/mcp.M900114-MCP200
In vivo stable isotope labeling of fruit flies reveals post-transcriptional regulation in the maternal-to-zygotic transition
Abstract
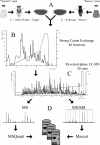
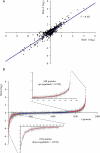
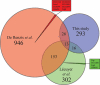
An important hallmark in embryonic development is characterized by the maternal-to-zygotic transition (MZT) where zygotic transcription is activated by a maternally controlled environment. Post-transcriptional and translational regulation is critical for this transition and has been investigated in considerable detail at the gene level. We used a proteomics approach using metabolic labeling of Drosophila to quantitatively assess changes in protein expression levels before and after the MZT. By combining stable isotope labeling of fruit flies in vivo with high accuracy quantitative mass spectrometry we could quantify 2,232 proteins of which about half changed in abundance during this process. We show that approximately 500 proteins increased in abundance, providing direct evidence of the identity of proteins as a product of embryonic translation. The group of down-regulated proteins is dominated by maternal factors involved in translational control of maternal and zygotic transcripts. Surprisingly a direct comparison of transcript and protein levels showed that the mRNA levels of down-regulated proteins remained relatively constant, indicating a translational control mechanism specifically targeting these proteins. In addition, we found evidence for post-translational processing of cysteine proteinase-1 (Cathepsin L), which became activated during the MZT as evidenced by the loss of its N-terminal propeptide. Poly(A)-binding protein was shown to be processed at its C-terminal tail, thereby losing one of its protein-interacting domains. Altogether this quantitative proteomics study provides a dynamic profile of known and novel proteins of maternal as well as embryonic origin. This provides insight into the production, stability, and modification of individual proteins, whereas discrepancies between transcriptional profiles and protein dynamics indicate novel control mechanisms in genome activation during early fly development.
Figures


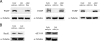
References
-
- Wieschaus E., Gehring W.( 1976) Clonal analysis of primordial disc cells in the early embryo of Drosophila melanogaster. Dev. Biol. 50, 249– 263 - PubMed
-
- Zalokar M.( 1976) Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev. Biol. 49, 425– 437 - PubMed
-
- Anderson K. V., Lengyel J. A.( 1979) Rates of synthesis of major classes of RNA in Drosophila embryos. Dev. Biol. 70, 217– 231 - PubMed
-
- Newport J., Kirschner M.( 1982) A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675– 686 - PubMed
-
- Edgar B. A., Schubiger G.( 1986) Parameters controlling transcriptional activation during early Drosophila development. Cell 44, 871– 877 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

