Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions
- PMID: 19321500
- PMCID: PMC2691829
- DOI: 10.1093/nar/gkp190
Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions
Abstract
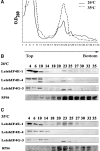
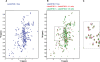
Translation initiation in eukaryotes is mediated by assembly of the eIF4F complex over the m(7)GTP cap structure at the 5'-end of mRNAs. This requires an interaction between eIF4E and eIF4G, two eIF4F subunits. The Leishmania orthologs of eIF4E are structurally diverged from their higher eukaryote counterparts, since they have evolved to bind the unique trypanosomatid cap-4 structure. Here, we characterize a key eIF4G candidate from Leishmania parasites (LeishIF4G-3) that contains a conserved MIF4G domain. LeishIF4G-3 was found to coelute with the parasite eIF4F subunits from an m(7)GTP-Sepharose column and to bind directly to LeishIF4E. In higher eukaryotes the eIF4E-eIF4G interaction is based on a conserved peptide signature [Y(X(4))Lphi], where X is any amino acid and Phi is a hydrophobic residue. A parallel eIF4E-binding peptide was identified in LeishIF4G-3 (20-YPGFSLDE-27). However, the binding motif varies extensively: in addition to Y20 and L25, binding strictly requires the presence of F23, whereas the hydrophobic amino acid (Phi) is dispensable. The LeishIF4E-LeishIF4G-3 interaction was also confirmed by nuclear magnetic resonance (NMR) studies. In view of these diversities, the characterization of the parasite eIF4E-eIF4G interaction may not only serve as a novel target for inhibiting Leishmaniasis but also provide important insight for future drug discovery.
Figures



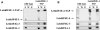


References
-
- Zilberstein D, Shapira M. The role of pH and temperature on the development of Leishmania parasites. Annu. Rev. Microbiol. 1994;48:449–470. - PubMed
-
- Bates P. Axenic culture of Leishmania amastigotes. Parasitol. Today. 1993;9:143–146. - PubMed
-
- Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, Zilberstein D. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol. Biochem. Parasitol. 2005;141:99–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

