New insights from old bones: DNA preservation and degradation in permafrost preserved mammoth remains
- PMID: 19321502
- PMCID: PMC2691819
- DOI: 10.1093/nar/gkp159
New insights from old bones: DNA preservation and degradation in permafrost preserved mammoth remains
Abstract
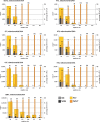
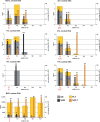
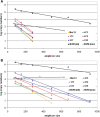
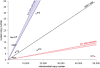
Despite being plagued by heavily degraded DNA in palaeontological remains, most studies addressing the state of DNA degradation have been limited to types of damage which do not pose a hindrance to Taq polymerase during PCR. Application of serial qPCR to the two fractions obtained during extraction (demineralization and protein digest) from six permafrost mammoth bones and one partially degraded modern elephant bone has enabled further insight into the changes which endogenous DNA is subjected to during diagenesis. We show here that both fractions exhibit individual qualities in terms of the prevailing type of DNA (i.e. mitochondrial versus nuclear DNA) as well as the extent of damage, and in addition observed a highly variable ratio of mitochondrial to nuclear DNA among the six mammoth samples. While there is evidence suggesting that mitochondrial DNA is better preserved than nuclear DNA in ancient permafrost samples, we find the initial DNA concentration in the bone tissue to be as relevant for the total accessible mitochondrial DNA as the extent of DNA degradation post-mortem. We also evaluate the general applicability of indirect measures of preservation such as amino-acid racemization, bone crystallinity index and thermal age to these exceptionally well-preserved samples.
Figures
References
-
- Debruyne R, Schwarz C, Poinar H. Comment on “Whole-Genome Shotgun Sequencing of Mitochondria from Ancient Hair Shafts”. Science. 2008;322:857. - PubMed
-
- Paabo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004;38:645–679. - PubMed
-
- Shemesh A. Crystallinity and diagenesis of sedimentary apatites. Geochim. Cosmochim. Acta. 1990;54:2433–2438.
-
- Poinar HN, Hoss M, Bada JL, Paabo S. Amino acid racemization and the preservation of ancient DNA. Science. 1996;272:864–866. - PubMed

