Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma
- PMID: 19321746
- PMCID: PMC2667034
- DOI: 10.1073/pnas.0900912106
Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma
Abstract
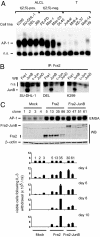
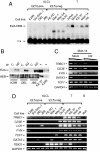
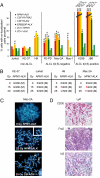
Although the identification and characterization of translocations have rapidly increased, little is known about the mechanisms of how translocations occur in vivo. We used anaplastic large cell lymphoma (ALCL) with and without the characteristic t(2;5)(p23;q35) translocation to study the mechanisms of formation of translocations and of ALCL transformation. We report deregulation of several genes located near the ALCL translocation breakpoint, regardless of whether the tumor contains the t(2;5). The affected genes include the oncogenic transcription factor Fra2 (located on 2p23), the HLH protein Id2 (2p25), and the oncogenic tyrosine kinase CSF1-receptor (5q33.1). Their up-regulation promotes cell survival and repression of T cell-specific gene expression programs that are characteristic for ALCL. The deregulated genes are in spatial proximity within the nuclear space of t(2;5)-negative ALCL cells, facilitating their translocation on induction of double-strand breaks. These data suggest that deregulation of breakpoint-proximal genes occurs before the formation of translocations, and that aberrant transcriptional activity of genomic regions is linked to their propensity to undergo chromosomal translocations. Also, our data demonstrate that deregulation of breakpoint-proximal genes has a key role in ALCL.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. - PubMed
-
- Janz S, Potter M, Rabkin CS. Lymphoma- and leukemia-associated chromosomal translocations in healthy individuals. Genes Chromosomes Cancer. 2003;36:211–223. - PubMed
-
- Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. - PubMed
-
- Stein H, et al. CD30(+) anaplastic large cell lymphoma: A review of its histopathologic, genetic, and clinical features. Blood. 2000;96:3681–3695. - PubMed
-
- Mathas S, et al. Elevated NF-kappaB p50 complex formation and Bcl-3 expression in classical Hodgkin, anaplastic large-cell, and other peripheral T-cell lymphomas. Blood. 2005;106:4287–4293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

