A mobile kinesin-head intermediate during the ATP-waiting state
- PMID: 19321748
- PMCID: PMC2667011
- DOI: 10.1073/pnas.0808355106
A mobile kinesin-head intermediate during the ATP-waiting state
Abstract
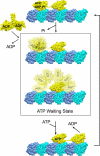
Kinesin1 is a motor protein that uses the energy from ATP hydrolysis to move intracellular cargoes along microtubules. It contains 2 identical motor domains, or heads, that coordinate their mechano-chemical cycles to move processively along microtubules. The molecular mechanism of coordination between head domains remains unclear, partly because of the lack of structural information on critical intermediates of the kinesin1 mechano-chemical cycle. A point of controversy has been whether before ATP binding, in the so called ATP-waiting state, 1 or 2 motor domains are bound to the microtubule. To address this issue, here we use ensemble and single molecule fluorescence polarization microscopy (FPM) to determine the mobility and orientation of the kinesin1 heads at different ATP concentrations and in heterodimeric constructs with microtubule binding impaired in 1 head. We found evidence for a mobile head during the ATP-waiting state. We incorporate our results into a model for kinesin translocation that accounts well for many reported experimental results.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Endow SA. Kinesin motors as molecular machines. Bioessays. 2003;25:1212–1219. - PubMed
-
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. - PubMed
-
- Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. - PubMed
-
- Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–393. - PubMed
-
- Coy DL, Wagenbach M, Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem. 1999;274:3667–3671. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

