A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis
- PMID: 19321749
- PMCID: PMC2667043
- DOI: 10.1073/pnas.0902019106
A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis
Abstract
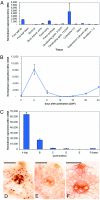
The walls of grasses and related members of the Poales are characterized by the presence of the polysaccharide (1,3, 1,4)-beta-D-glucan (beta-glucan). To date, only members of the grass-specific cellulose synthase-like F (CSLF) gene family have been implicated in its synthesis. Assuming that other grass-specific CSL genes also might encode synthases for this polysaccharide, we cloned HvCSLH1, a CSLH gene from barley (Hordeum vulgare L.), and expressed an epitope-tagged version of the cDNA in Arabidopsis, a species with no CSLH genes and no beta-glucan in its walls. Transgenic Arabidopsis lines that had detectable amounts of the epitope-tagged HvCSLH1 protein accumulated beta-glucan in their walls. The presence of beta-glucan was confirmed by immunoelectron microscopy (immuno-EM) of sectioned tissues and chemical analysis of wall extracts. In the chemical analysis, characteristic tri- and tetra-saccharides were identified by high-performance anion-exchange chromatography and MALDI-TOF MS following their release from transgenic Arabidopsis walls by a specific beta-glucan hydrolase. Immuno-EM also was used to show that the epitope-tagged HvCSLH1 protein was in the endoplasmic reticulum and Golgi-associated vesicles, but not in the plasma membrane. In barley, HvCSLH1 was expressed at very low levels in leaf, floral tissues, and the developing grain. In leaf, expression was highest in xylem and interfascicular fiber cells that have walls with secondary thickenings containing beta-glucan. Thus both the CSLH and CSLF families contribute to beta-glucan synthesis in grasses and probably do so independently of each other, because there is no significant transcriptional correlation between these genes in the barley tissues surveyed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Genetics and physiology of cell wall polysaccharides in the model C4 grass, Setaria viridis spp.BMC Plant Biol. 2015 Oct 2;15:236. doi: 10.1186/s12870-015-0624-0. BMC Plant Biol. 2015. PMID: 26432387 Free PMC article.
-
Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure.Plant Biotechnol J. 2011 Feb;9(2):117-35. doi: 10.1111/j.1467-7652.2010.00532.x. Plant Biotechnol J. 2011. PMID: 20497371
-
Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans.Science. 2006 Mar 31;311(5769):1940-2. doi: 10.1126/science.1122975. Science. 2006. PMID: 16574868
-
(1,3;1,4)-beta-D-glucans in cell walls of the poaceae, lower plants, and fungi: a tale of two linkages.Mol Plant. 2009 Sep;2(5):873-82. doi: 10.1093/mp/ssp063. Epub 2009 Aug 17. Mol Plant. 2009. PMID: 19825664 Review.
-
Polysaccharide hydrolases in germinated barley and their role in the depolymerization of plant and fungal cell walls.Int J Biol Macromol. 1997 Aug;21(1-2):67-72. doi: 10.1016/s0141-8130(97)00043-3. Int J Biol Macromol. 1997. PMID: 9283018 Review.
Cited by
-
Expression analysis of cellulose synthase-like genes in durum wheat.Sci Rep. 2018 Oct 23;8(1):15675. doi: 10.1038/s41598-018-34013-6. Sci Rep. 2018. PMID: 30353138 Free PMC article.
-
Differences in protein structural regions that impact functional specificity in GT2 family β-glucan synthases.PLoS One. 2019 Oct 30;14(10):e0224442. doi: 10.1371/journal.pone.0224442. eCollection 2019. PLoS One. 2019. PMID: 31665152 Free PMC article.
-
Biosynthesis and Transport of Nucleotide Sugars for Plant Hemicellulose.Front Plant Sci. 2021 Nov 12;12:723128. doi: 10.3389/fpls.2021.723128. eCollection 2021. Front Plant Sci. 2021. PMID: 34868108 Free PMC article. Review.
-
Species-Specific Gene Expansion of the Cellulose synthase Gene Superfamily in the Orchidaceae Family and Functional Divergence of Mannan Synthesis-Related Genes in Dendrobium officinale.Front Plant Sci. 2022 Jun 3;13:777332. doi: 10.3389/fpls.2022.777332. eCollection 2022. Front Plant Sci. 2022. PMID: 35720557 Free PMC article.
-
Evolution, gene expression profiling and 3D modeling of CSLD proteins in cotton.BMC Plant Biol. 2017 Jul 10;17(1):119. doi: 10.1186/s12870-017-1063-x. BMC Plant Biol. 2017. PMID: 28693426 Free PMC article.
References
-
- Harris PJ. Diversity in plant cell walls. In: Henry RJ, editor. Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants. Wallingford, Oxon, UK: CAB International Publishing; 2005. pp. 201–227.
-
- Trethewey JAK, Campbell LM, Harris PJ. (1→3),(1→4)-β-D-Glucans in the cell walls of the Poales (sensu lato): An immunogold labeling study using a monoclonal antibody. Am J Bot. 2005;92:1660–1674. - PubMed
-
- Carpita NC. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. - PubMed
-
- Trethewey JAK, Harris PJ. Location of (1→3),(1→4)-β-D-glucans in vegetative cell walls of barley (Hordeum vulgare) using immunogold labelling. New Phytol. 2002;154:347–358. - PubMed
-
- Fincher GB, Stone BA. Chemistry of nonstarch polysaccharides. In: Wrigley C, Corke H, Walker CE, editors. Encyclopedia of Grain Science. Vol 1. London: Academic Press; 2004. pp. 206–222.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

