A large-scale international meta-analysis of paraoxonase gene polymorphisms in sporadic ALS
- PMID: 19321847
- PMCID: PMC2707108
- DOI: 10.1212/WNL.0b013e3181a18674
A large-scale international meta-analysis of paraoxonase gene polymorphisms in sporadic ALS
Abstract
Background: Six candidate gene studies report a genetic association of DNA variants within the paraoxonase locus with sporadic amyotrophic lateral sclerosis (ALS). However, several other large studies, including five genome-wide association studies, have not duplicated this finding.
Methods: We conducted a meta-analysis of 10 published studies and one unpublished study of the paraoxonase locus, encompassing 4,037 ALS cases and 4,609 controls, including genome-wide association data from 2,018 ALS cases and 2,425 controls.
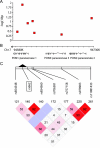
Results: The combined fixed effects odds ratio (OR) for rs662 (PON1 Q192R) was 1.09 (95% confidence interval [CI], 1.02-1.16, p = 0.01); the genotypic OR for RR homozygotes at Q192R was 1.25 (95% CI, 1.07-1.45, p = 0.0004); the combined OR for rs854560 (PON1 L55M) was 0.97 (95% CI, 0.86-1.10, p = 0.62); the OR for rs10487132 (PON2) was 1.08 (95% CI, 0.92-1.27, p = 0.35). Although the rs662 polymorphism reached a nominal level of significance, no polymorphism was significant after multiple testing correction. In the subanalysis of samples with genome-wide data from which population outliers were removed, rs662 had an OR of 1.06 (95% CI, 0.97-1.16, p = 0.22).
Conclusions: In contrast to previous positive smaller studies, our genetic meta-analysis showed no significant association of amyotrophic lateral sclerosis (ALS) with the PON locus. This is the largest meta-analysis of a candidate gene in ALS to date and the first ALS meta-analysis to include data from whole genome association studies. The findings reinforce the need for much larger and more collaborative investigations of the genetic determinants of ALS.
Figures

Comment in
-
Paraoxonase genes and susceptibility to ALS.Neurology. 2009 Jul 7;73(1):11-2. doi: 10.1212/WNL.0b013e3181aa2a39. Epub 2009 May 13. Neurology. 2009. PMID: 19439719 No abstract available.
References
-
- Schymick JC, Talbot K, Traynor BJ. Genetics of sporadic amyotrophic lateral sclerosis. Hum Mol Genet 2007;16:R233–242. - PubMed
-
- Simpson CL, Al-Chalabi A. Amyotrophic lateral sclerosis as a complex genetic disease. Biochim Biophys Acta 2006;1762:973–985. - PubMed
-
- Slowik A, Tomik B, Wolkow PP, et al. Paraoxonase gene polymorphisms and sporadic ALS. Neurology 2006;67:766–770. - PubMed
-
- Saeed M, Siddique N, Hung WY, et al. Paraoxonase cluster polymorphisms are associated with sporadic ALS. Neurology 2006;67:771–776. - PubMed
-
- Morahan JM, Yu B, Trent RJ, Pamphlett R. A gene-environment study of the paraoxonase 1 gene and pesticides in amyotrophic lateral sclerosis. Neurotoxicology 2007;28:532–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
