Efficient control of group I intron ribozyme catalysis by DNA constraints
- PMID: 19322435
- PMCID: PMC2662395
- DOI: 10.1039/b820676g
Efficient control of group I intron ribozyme catalysis by DNA constraints
Abstract
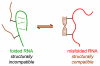
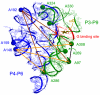
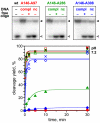
Double-stranded DNA constraints enable efficient control of catalysis by a large multi-domain group I intron ribozyme.
Figures
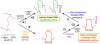
Similar articles
-
A role for a single-stranded junction in RNA binding and specificity by the Tetrahymena group I ribozyme.J Am Chem Soc. 2012 Feb 1;134(4):1910-3. doi: 10.1021/ja2083575. Epub 2012 Jan 17. J Am Chem Soc. 2012. PMID: 22220837 Free PMC article.
-
Biogenic triamine and tetraamine activate core catalytic ability of Tetrahymena group I ribozyme in the absence of its large activator module.Biochem Biophys Res Commun. 2018 Feb 5;496(2):594-600. doi: 10.1016/j.bbrc.2018.01.085. Epub 2018 Jan 12. Biochem Biophys Res Commun. 2018. PMID: 29339152
-
Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site.Mol Cell. 2004 Nov 5;16(3):351-62. doi: 10.1016/j.molcel.2004.10.003. Mol Cell. 2004. PMID: 15525509
-
The structure and function of catalytic RNAs.Sci China C Life Sci. 2009 Mar;52(3):232-44. doi: 10.1007/s11427-009-0038-z. Epub 2009 Mar 18. Sci China C Life Sci. 2009. PMID: 19294348 Review.
-
Maximizing RNA folding rates: a balancing act.RNA. 2000 Jun;6(6):790-4. doi: 10.1017/s1355838200000522. RNA. 2000. PMID: 10864039 Free PMC article. Review.
Cited by
-
Mg2+-dependent conformational changes and product release during DNA-catalyzed RNA ligation monitored by Bimane fluorescence.Nucleic Acids Res. 2015 Jan;43(1):40-50. doi: 10.1093/nar/gku1268. Epub 2014 Dec 10. Nucleic Acids Res. 2015. PMID: 25505142 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

