Functionally important conformations of the Met20 loop in dihydrofolate reductase are populated by rapid thermal fluctuations
- PMID: 19323547
- PMCID: PMC2889193
- DOI: 10.1021/ja9000135
Functionally important conformations of the Met20 loop in dihydrofolate reductase are populated by rapid thermal fluctuations
Abstract
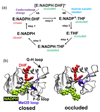
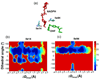
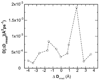
Conformational changes in enzymes are well recognized to play an important role in the organization of the reactive groups for efficient catalysis. This study reveals atomic and energetic details of the conformational change process that precedes the catalytic reaction of the enzyme dihydrofolate reductase. The computed free energy profile provides insights into the ligand binding mechanism and a quantitative estimate of barrier heights separating different conformational states along the pathway. Studies show that the ternary complex comprised of NADPH cofactor and substrate dihydrofolate undergoes transitions between a closed state and an occluded state via an intermediate "open" conformation. During these transitions the largest conformational change occurs in the Met20 loop of DHFR and is accompanied by the motion of the cofactor into and out of the binding pocket. When the cofactor is out of the binding pocket, the enzyme frequently samples open and occluded conformations with a small (approximately 5 k(B)T) free energy barrier between the two states. However, when the cofactor is in the binding pocket, the closed conformation is thermodynamically most favored. The determination of a profile characterizing the position-dependent diffusion of the Met20 loop allowed us to apply reaction rate theory and deduce the kinetics of loop motions based on the computed free energy landscape.
Figures


References
-
- Blakely RL. In: Chemistry and Biochemistry of Folates. In folates and pterins. Blakely RL, Benkovic SJ, editors. Vol. 3. New York: Wiley; 1984. pp. 191–253.
-
- Radkiewicz JL, Brooks CL., III J. Am. Chem. Soc. 2000;122:225–231.
- Agarwal PK, Billeter SR, Rajagopalan PTR, Benkovic SJ, Hammes-Schiffer S. Proc. Nat. Acad. Sci. USA. 2002;99:2794–2799. - PMC - PubMed
- Rod TH, Brooks CL., III J. Am. Chem. Soc. 2003;125:8718–8719. - PubMed
- Rod TH, Radkiewicz JL, Brooks CL., III Proc. Nat. Acad. Sci. USA. 2003;100:6980–6985. - PMC - PubMed
- Thorpe IF, Brooks CL., III J. Phys. Chem. B. 2003;107:14042–14051.
- Thorpe IF, Brooks CL., III Proteins: Struc. Func. Gen. 2004;57:444–457. - PubMed
- Thorpe IF, Brooks CL., III J. Amer. Chem. Soc. 2005;127:12997–13006. - PubMed
- Okazaki K, Koga N, Takada S, Onuchic JN, Wolynes PG. Proc Natl Acad Sci U S A. 2006;103:11844–11849. - PMC - PubMed
- Chen J, Dima RI, Thirumalai D. J Mol Biol. 2007;374:250–266. - PubMed
-
- Fierke CA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:4085–4092. - PubMed
- Epstein DM, Benkovic SJ, Wright PE. Biochemistry. 1995;34:11037–11048. - PubMed
- Cannon WR, Singleton SE, Benkovic SJ. Nat. Struct. Biol. 1996;3:821–833. - PubMed
- Miller GP, Benkovic SJ. Biochemistry. 1998;37:6327–6335. - PubMed
- Miller GP, Wahnon DC, Benkovic SJ. Biochemistry. 2001;40:867–875. - PubMed
- Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Biochemistry. 2001;40:9846–9859. - PubMed
- Rajagopalan PTR, Lutz S, Benkovic SJ. Biochemistry. 2002;41:12618–12628. - PubMed
- Osborne MJ, Venkitakrishnan RP, Dyson HJ, Wright PE. Protein Sci. 2003;12:2230–2238. - PMC - PubMed
- Schnell JR, Dyson HJ, Wright PE. Biochemistry. 2004;43:374–383. - PubMed
- Venkitakrishnan RP, Zaborowski E, McElheny D, Benkovic SJ, Dyson HJ, Wright PE. Biochemistry. 2004;43:16046–16055. - PubMed
- McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Proc. Nat. Acad. Sci. USA. 2005;102:5032–5037. - PMC - PubMed
- Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638–1642. - PubMed
- Boehr DD, Dyson HJ, Wright PE. Biochemistry. 2008;47:9227–9233. - PMC - PubMed
-
- Sawaya MR, Kraut J. Biochemistry. 1997;36:586–603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

