Challenges in the computational design of proteins
- PMID: 19324680
- PMCID: PMC2843960
- DOI: 10.1098/rsif.2008.0508.focus
Challenges in the computational design of proteins
Abstract
Protein design has many applications not only in biotechnology but also in basic science. It uses our current knowledge in structural biology to predict, by computer simulations, an amino acid sequence that would produce a protein with targeted properties. As in other examples of synthetic biology, this approach allows the testing of many hypotheses in biology. The recent development of automated computational methods to design proteins has enabled proteins to be designed that are very different from any known ones. Moreover, some of those methods mostly rely on a physical description of atomic interactions, which allows the designed sequences not to be biased towards known proteins. In this paper, we will describe the use of energy functions in computational protein design, the use of atomic models to evaluate the free energy in the unfolded and folded states, the exploration and optimization of amino acid sequences, the problem of negative design and the design of biomolecular function. We will also consider its use together with the experimental techniques such as directed evolution. We will end by discussing the challenges ahead in computational protein design and some of their future applications.
Figures

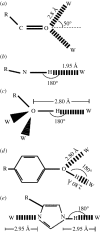


 and
and  .
.  reflects the probability of the ligand binding and not inducing the corresponding conformational change.
reflects the probability of the ligand binding and not inducing the corresponding conformational change.  reflects the probability of the conformational transition taking place in the absence of the ligand. The probability that a designed protein will adopt the target state is given by
reflects the probability of the conformational transition taking place in the absence of the ligand. The probability that a designed protein will adopt the target state is given by  .
.

References
-
- Antonkine M. L., Maes E. M., Czernuszewicz R. S., Breitenstein E. B., Falzone C. J., Balasubramanian R., Lubner C., Bryant D. A., Golbeck J. H. 2007. Chemical rescue of a site-modified ligand to a [4Fe–4S] cluster in a bacterial di-cluster ferredoxin. Biochim. Biophys. Acta. 1767, 712–724. (10.1016/j.bbabio.2007.02.003) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
