Genetic modulation of energy metabolism in birds through mitochondrial function
- PMID: 19324832
- PMCID: PMC2660998
- DOI: 10.1098/rspb.2008.1946
Genetic modulation of energy metabolism in birds through mitochondrial function
Abstract
Despite their central importance for the evolution of physiological variation, the genetic mechanisms that determine energy expenditure in animals have largely remained unstudied. We used quantitative genetics to confirm that both mass-specific and whole-organism basal metabolic rate (BMR) were heritable in a captive-bred population of stonechats (Saxicola torquata spp.) founded on birds from three wild populations (Europe, Africa and Asia) that differed in BMR. This argues that BMR is at least partially under genetic control by multiple unknown nuclear loci each with a limited effect on the phenotype. We then tested for a genetic effect on BMR based on mitochondrial-nuclear coadaptation using hybrids between ancestral populations with high and low BMR (Europe-Africa and Asia-Europe), with different parental configurations (female(high)-male(low) or female(low)-male(high)) within each combination of populations. Hybrids with different parental configurations have on average identical mixtures of nuclear DNA, but differ in mitochondrial DNA because it is inherited only from the mother. Mass-specific BMR differed between hybrids with different parental configurations, implying that the combination of mitochondrial and nuclear DNA affected metabolic rate. Therefore, our findings implicate mitochondrial function as an important regulator of energy metabolism. In combination with the substantial heritabilities of metabolic rate, and corroborated by genetic differences in the mitochondrial genome, these results set the stage for further investigations of a genetic control mechanism involving both mitochondrial and nuclear genes determining metabolic rate at the whole-organism level.
Figures
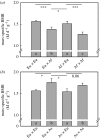
References
-
- Avise J.C., Nelson W.S. Molecular genetic relationships of the extinct dusky seaside sparrow. Science. 1989;243:646–648. doi:10.1126/science.243.4891.646 - DOI - PubMed
-
- Bai Y.D., Shakeley R.M., Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell. Biol. 2000;20:805–815. doi:10.1128/MCB.20.3.805-815.2000 - DOI - PMC - PubMed
-
- Ballard J.W.O., Rand D.M. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 2005;36:621–642. doi:10.1146/annurev.ecolsys.36.091704.175513 - DOI
-
- Ballard J.W.O., Whitlock M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi:10.1046/j.1365-294X.2003.02063.x - DOI - PubMed
-
- Blier P.U., Dufresne F., Burton R.S. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 2001;17:400–406. doi:10.1016/S0168-9525(01)02338-1 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

